SARS-CoV-2 cellular tropism and direct multiorgan failure in COVID-19 patients: Bioinformatic predictions, experimental observations, and open questions
- PMID: 36229927
- PMCID: PMC9874490
- DOI: 10.1002/cbin.11928
SARS-CoV-2 cellular tropism and direct multiorgan failure in COVID-19 patients: Bioinformatic predictions, experimental observations, and open questions
Abstract
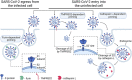
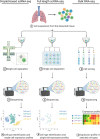
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19), has led to an unprecedented public health emergency worldwide. While common cold symptoms are observed in mild cases, COVID-19 is accompanied by multiorgan failure in severe patients. Organ damage in COVID-19 patients is partially associated with the indirect effects of SARS-CoV-2 infection (e.g., systemic inflammation, hypoxic-ischemic damage, coagulopathy), but early processes in COVID-19 patients that trigger a chain of indirect effects are connected with the direct infection of cells by the virus. To understand the virus transmission routes and the reasons for the wide-spectrum of complications and severe outcomes of COVID-19, it is important to identify the cells targeted by SARS-CoV-2. This review summarizes the major steps of investigation and the most recent findings regarding SARS-CoV-2 cellular tropism and the possible connection between the early stages of infection and multiorgan failure in COVID-19. The SARS-CoV-2 pandemic is the first epidemic in which data extracted from single-cell RNA-seq (scRNA-seq) gene expression data sets have been widely used to predict cellular tropism. The analysis presented here indicates that the SARS-CoV-2 cellular tropism predictions are accurate enough for estimating the potential susceptibility of different cells to SARS-CoV-2 infection; however, it appears that not all susceptible cells may be infected in patients with COVID-19.
Keywords: SARS-CoV-2; cell entry factors; cellular tropism; infection; multiorgan failure; scRNA-seq.
© 2022 International Federation of Cell Biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study.Lancet Microbe. 2020 May;1(1):e14-e23. doi: 10.1016/S2666-5247(20)30004-5. Epub 2020 Apr 21. Lancet Microbe. 2020. PMID: 32835326 Free PMC article.
-
Identification of Transcription Factors Regulating SARS-CoV-2 Tropism Factor Expression by Inferring Cell-Type-Specific Transcriptional Regulatory Networks in Human Lungs.Viruses. 2022 Apr 17;14(4):837. doi: 10.3390/v14040837. Viruses. 2022. PMID: 35458567 Free PMC article.
-
Evidence of Severe Acute Respiratory Syndrome Coronavirus 2 Replication and Tropism in the Lungs, Airways, and Vascular Endothelium of Patients With Fatal Coronavirus Disease 2019: An Autopsy Case Series.J Infect Dis. 2021 Mar 3;223(5):752-764. doi: 10.1093/infdis/jiab039. J Infect Dis. 2021. PMID: 33502471 Free PMC article.
-
Molecular mechanisms implicated in SARS-CoV-2 liver tropism.World J Gastroenterol. 2022 Dec 28;28(48):6875-6887. doi: 10.3748/wjg.v28.i48.6875. World J Gastroenterol. 2022. PMID: 36632318 Free PMC article. Review.
-
Cardiovascular Tropism and Sequelae of SARS-CoV-2 Infection.Viruses. 2022 May 25;14(6):1137. doi: 10.3390/v14061137. Viruses. 2022. PMID: 35746609 Free PMC article. Review.
Cited by
-
SARS-Cov-2 Replication in a Blood-Brain Barrier Model Established with Human Brain Microvascular Endothelial Cells Induces Permeability and Disables ACE2-Dependent Regulation of Bradykinin B1 Receptor.Int J Mol Sci. 2025 Jun 10;26(12):5540. doi: 10.3390/ijms26125540. Int J Mol Sci. 2025. PMID: 40565006 Free PMC article.
-
SARS-CoV-2 mechanisms of cell tropism in various organs considering host factors.Heliyon. 2024 Feb 20;10(4):e26577. doi: 10.1016/j.heliyon.2024.e26577. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38420467 Free PMC article. Review.
-
The Nucleocapsid (N) Proteins of Different Human Coronaviruses Demonstrate a Variable Capacity to Induce the Formation of Cytoplasmic Condensates.Int J Mol Sci. 2024 Dec 7;25(23):13162. doi: 10.3390/ijms252313162. Int J Mol Sci. 2024. PMID: 39684875 Free PMC article.
-
Vascular dysfunction in hemorrhagic viral fevers: opportunities for organotypic modeling.Biofabrication. 2024 Jun 5;16(3):032008. doi: 10.1088/1758-5090/ad4c0b. Biofabrication. 2024. PMID: 38749416 Free PMC article. Review.
-
Zebrafish models of COVID-19.FEMS Microbiol Rev. 2023 Jan 16;47(1):fuac042. doi: 10.1093/femsre/fuac042. FEMS Microbiol Rev. 2023. PMID: 36323404 Free PMC article. Review.
References
-
- Acheampong, K. K. , Schaff, D. L. , Emert, B. L. , Lake, J. , Reffsin, S. , Shea, E. K. , Comar, C. E. , Litzky, L. A. , Khurram, N. A. , Linn, R. L. , Feldman, M. , Weiss, S. R. , Montone, K. T. , Cherry, S. , & Shaffer, S. M. (2022). Subcellular detection of SARS‐CoV‐2 RNA in human tissue reveals distinct localization in alveolar type 2 pneumocytes and alveolar macrophages. mBio, 13, e0375121. 10.1128/mbio.03751-21 - DOI - PMC - PubMed
-
- Ahn, J. H. , Kim, J. , Hong, S. P. , Choi, S. Y. , Yang, M. J. , Ju, Y. S. , Kim, Y. T. , Kim, H. M. , Rahman, M. T. , Chung, M. K. , Hong, S. D. , Bae, H. , Lee, C. S. , & Koh, G. Y. (2021). Nasal ciliated cells are primary targets for SARS‐CoV‐2 replication in the early stage of COVID‐19. Journal of Clinical Investigation, 131(13), e148517. 10.1172/JCI148517 - DOI - PMC - PubMed
-
- Amraei, R. , Yin, W. , Napoleon, M. A. , Suder, E. L. , Berrigan, J. , Zhao, Q. , Olejnik, J. , Chandler, K. B. , Xia, C. , Feldman, J. , Hauser, B. M. , Caradonna, T. M. , Schmidt, A. G. , Gummuluru, S. , Mühlberger, E. , Chitalia, V. , Costello, C. E. , & Rahimi, N. (2021). CD209L/L‐SIGN and CD209/DC‐SIGN act as receptors for SARS‐CoV‐2. ACS Central Science, 7(7), 1156–1165. 10.1021/acscentsci.0c01537 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

