Machine Learning-Based Genome-Wide Salivary DNA Methylation Analysis for Identification of Noninvasive Biomarkers in Oral Cancer Diagnosis
- PMID: 36230858
- PMCID: PMC9563273
- DOI: 10.3390/cancers14194935
Machine Learning-Based Genome-Wide Salivary DNA Methylation Analysis for Identification of Noninvasive Biomarkers in Oral Cancer Diagnosis
Abstract
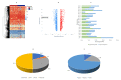
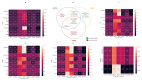
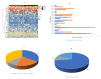
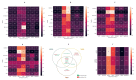
This study aims to examine the feasibility of ML-assisted salivary-liquid-biopsy platforms using genome-wide methylation analysis at the base-pair and regional resolution for delineating oral squamous cell carcinoma (OSCC) and oral potentially malignant disorders (OPMDs). A nested cohort of patients with OSCC and OPMDs was randomly selected from among patients with oral mucosal diseases. Saliva samples were collected, and DNA extracted from cell pellets was processed for reduced-representation bisulfite sequencing. Reads with a minimum of 10× coverage were used to identify differentially methylated CpG sites (DMCs) and 100 bp regions (DMRs). The performance of eight ML models and three feature-selection methods (ANOVA, MRMR, and LASSO) were then compared to determine the optimal biomarker models based on DMCs and DMRs. A total of 1745 DMCs and 105 DMRs were identified for detecting OSCC. The proportion of hypomethylated and hypermethylated DMCs was similar (51% vs. 49%), while most DMRs were hypermethylated (62.9%). Furthermore, more DMRs than DMCs were annotated to promoter regions (36% vs. 16%) and more DMCs than DMRs were annotated to intergenic regions (50% vs. 36%). Of all the ML models compared, the linear SVM model based on 11 optimal DMRs selected by LASSO had a perfect AUC, recall, specificity, and calibration (1.00) for OSCC detection. Overall, genome-wide DNA methylation techniques can be applied directly to saliva samples for biomarker discovery and ML-based platforms may be useful in stratifying OSCC during disease screening and monitoring.
Keywords: DNA methylation; biomarkers; diagnosis; epigenomics; oral cancer; oral potentially malignant disorders.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification of Differentially Methylated Sites with Weak Methylation Effects.Genes (Basel). 2018 Feb 8;9(2):75. doi: 10.3390/genes9020075. Genes (Basel). 2018. PMID: 29419727 Free PMC article.
-
Aberrant signature methylome by DNMT1 hot spot mutation in hereditary sensory and autonomic neuropathy 1E.Epigenetics. 2014 Aug;9(8):1184-93. doi: 10.4161/epi.29676. Epub 2014 Jul 7. Epigenetics. 2014. PMID: 25033457 Free PMC article.
-
Integrated DNA methylation analysis of peripheral blood from asbestos exposed populations and patients with malignant mesothelioma reveals novel methylation driver genes of diagnostic and prognostic relevance.Environ Pollut. 2024 Dec 1;362:124928. doi: 10.1016/j.envpol.2024.124928. Epub 2024 Sep 13. Environ Pollut. 2024. PMID: 39265763
-
Genome-wide DNA methylation profile identified a unique set of differentially methylated immune genes in oral squamous cell carcinoma patients in India.Clin Epigenetics. 2017 Feb 3;9:13. doi: 10.1186/s13148-017-0314-x. eCollection 2017. Clin Epigenetics. 2017. PMID: 28174608 Free PMC article.
-
Identification of regulatory role of DNA methylation in colon cancer gene expression via systematic bioinformatics analysis.Medicine (Baltimore). 2017 Nov;96(47):e8487. doi: 10.1097/MD.0000000000008487. Medicine (Baltimore). 2017. PMID: 29381923 Free PMC article.
Cited by
-
Methods in DNA methylation array dataset analysis: A review.Comput Struct Biotechnol J. 2024 May 17;23:2304-2325. doi: 10.1016/j.csbj.2024.05.015. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38845821 Free PMC article. Review.
-
Melting curve analyses in the quantitative real-time polymerase chain reaction of methylated/non-methylated DNA toward the detection of oral cancer using gargle fluid.Heliyon. 2025 Jan 24;11(3):e42286. doi: 10.1016/j.heliyon.2025.e42286. eCollection 2025 Feb 15. Heliyon. 2025. PMID: 39944336 Free PMC article.
-
RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma.Cancers (Basel). 2023 May 9;15(10):2672. doi: 10.3390/cancers15102672. Cancers (Basel). 2023. PMID: 37345009 Free PMC article.
-
Artificial Intelligence in Oral Cancer: A Comprehensive Scoping Review of Diagnostic and Prognostic Applications.Diagnostics (Basel). 2025 Jan 24;15(3):280. doi: 10.3390/diagnostics15030280. Diagnostics (Basel). 2025. PMID: 39941210 Free PMC article. Review.
-
Leveraging technology-driven strategies to untangle omics big data: circumventing roadblocks in clinical facets of oral cancer.Front Oncol. 2024 Jan 3;13:1183766. doi: 10.3389/fonc.2023.1183766. eCollection 2023. Front Oncol. 2024. PMID: 38234400 Free PMC article. Review.
References
-
- Adeoye J., Thomson P. Strategies to improve diagnosis and risk assessment for oral cancer patients. Fac. Dent. J. 2020;11:122–127. doi: 10.1308/rcsfdj.2020.97. - DOI
LinkOut - more resources
Full Text Sources

