Single Cell Transcriptomics to Understand HSC Heterogeneity and Its Evolution upon Aging
- PMID: 36231086
- PMCID: PMC9563410
- DOI: 10.3390/cells11193125
Single Cell Transcriptomics to Understand HSC Heterogeneity and Its Evolution upon Aging
Abstract
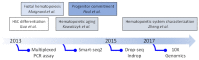
Single-cell transcriptomic technologies enable the uncovering and characterization of cellular heterogeneity and pave the way for studies aiming at understanding the origin and consequences of it. The hematopoietic system is in essence a very well adapted model system to benefit from this technological advance because it is characterized by different cellular states. Each cellular state, and its interconnection, may be defined by a specific location in the global transcriptional landscape sustained by a complex regulatory network. This transcriptomic signature is not fixed and evolved over time to give rise to less efficient hematopoietic stem cells (HSC), leading to a well-documented hematopoietic aging. Here, we review the advance of single-cell transcriptomic approaches for the understanding of HSC heterogeneity to grasp HSC deregulations upon aging. We also discuss the new bioinformatics tools developed for the analysis of the resulting large and complex datasets. Finally, since hematopoiesis is driven by fine-tuned and complex networks that must be interconnected to each other, we highlight how mathematical modeling is beneficial for doing such interconnection between multilayered information and to predict how HSC behave while aging.
Keywords: Boolean modeling; HSC aging; bioinformatics; single cell transcriptomic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

