Neomycin Interferes with Phosphatidylinositol-4,5-Bisphosphate at the Yeast Plasma Membrane and Activates the Cell Wall Integrity Pathway
- PMID: 36232332
- PMCID: PMC9569482
- DOI: 10.3390/ijms231911034
Neomycin Interferes with Phosphatidylinositol-4,5-Bisphosphate at the Yeast Plasma Membrane and Activates the Cell Wall Integrity Pathway
Abstract
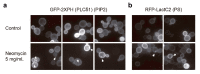
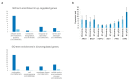
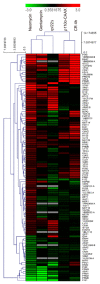
The cell wall integrity pathway (CWI) is a MAPK-mediated signaling route essential for yeast cell response to cell wall damage, regulating distinct aspects of fungal physiology. We have recently proven that the incorporation of a genetic circuit that operates as a signal amplifier into this pathway allows for the identification of novel elements involved in CWI signaling. Here, we show that the strong growth inhibition triggered by pathway hyperactivation in cells carrying the "Integrity Pathway Activation Circuit" (IPAC) also allows the easy identification of new stimuli. By using the IPAC, we have found various chemical agents that activate the CWI pathway, including the aminoglycoside neomycin. Cells lacking key components of this pathway are sensitive to this antibiotic, due to the disruption of signaling upon neomycin stimulation. Neomycin reduces both phosphatidylinositol-4,5-bisphosphate (PIP2) availability at the plasma membrane and myriocin-induced TORC2-dependent Ypk1 phosphorylation, suggesting a strong interference with plasma membrane homeostasis, specifically with PIP2. The neomycin-induced transcriptional profile involves not only genes related to stress and cell wall biogenesis, but also to amino acid metabolism, reflecting the action of this antibiotic on the yeast ribosome.
Keywords: MAPK; PIP2; Saccharomyces cerevisiae; Slt2; cell wall integrity; neomycin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

