Interspecies Comparison of Interaction Energies between Photosynthetic Protein RuBisCO and 2CABP Ligand
- PMID: 36232645
- PMCID: PMC9570433
- DOI: 10.3390/ijms231911347
Interspecies Comparison of Interaction Energies between Photosynthetic Protein RuBisCO and 2CABP Ligand
Abstract
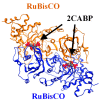
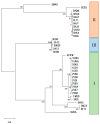
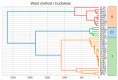
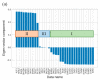
Ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) functions as the initial enzyme in the dark reactions of photosynthesis, catalyzing reactions that extract CO2 from the atmosphere and fix CO2 into organic compounds. RuBisCO is classified into four types (isoforms I-IV) according to sequence-based phylogenetic trees. Given its size, the computational cost of accurate quantum-chemical calculations for functional analysis of RuBisCO is high; however, recent advances in hardware performance and the use of the fragment molecular orbital (FMO) method have enabled the ab initio analyses of RuBisCO. Here, we performed FMO calculations on multiple structural datasets for various complexes with the 2'-carboxylarabinitol 1,5-bisphosphate (2CABP) ligand as a substrate analog and investigated whether phylogenetic relationships based on sequence information are physicochemically relevant as well as whether novel information unobtainable from sequence information can be revealed. We extracted features similar to the phylogenetic relationships found in sequence analysis, and in terms of singular value decomposition, we identified residues that strongly interacted with the ligand and the characteristics of the isoforms for each principal component. These results identified a strong correlation between phylogenetic relationships obtained by sequence analysis and residue interaction energies with the ligand. Notably, some important residues were located far from the ligand, making comparisons among species using only residues proximal to the ligand insufficient.
Keywords: fragment molecular orbital (FMO) method; inter-fragment interaction energy (IFIE); ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO); singular value decomposition (SVD).
Conflict of interest statement
The authors declare no conflict of interest.
Figures










References
-
- Matsumura H., Shiomi K., Yamamoto A., Taketani Y., Kobayashi N., Yoshizawa T., Tanaka S.I., Yoshikawa H., Endo M., Fukayama H. Hybrid Rubisco with complete replacement of rice Rubisco small by sorghum counterparts confers C4 plant-like high catalytic activity. Mol. Plant. 2020;13:1570–1581. doi: 10.1016/j.molp.2020.08.012. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

