SP1 and NFY Regulate the Expression of PNPT1, a Gene Encoding a Mitochondrial Protein Involved in Cancer
- PMID: 36232701
- PMCID: PMC9570217
- DOI: 10.3390/ijms231911399
SP1 and NFY Regulate the Expression of PNPT1, a Gene Encoding a Mitochondrial Protein Involved in Cancer
Abstract
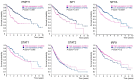
The Polyribonucleotide nucleotidyltransferase 1 gene (PNPT1) encodes polynucleotide phosphorylase (PNPase), a 3'-5' exoribonuclease involved in mitochondrial RNA degradation and surveillance and RNA import into the mitochondrion. Here, we have characterized the PNPT1 promoter by in silico analysis, luciferase reporter assays, electrophoretic mobility shift assays (EMSA), chromatin immunoprecipitation (ChIP), siRNA-based mRNA silencing and RT-qPCR. We show that the Specificity protein 1 (SP1) transcription factor and Nuclear transcription factor Y (NFY) bind the PNPT1 promoter, and have a relevant role regulating the promoter activity, PNPT1 expression, and mitochondrial activity. We also found in Kaplan-Meier survival curves that a high expression of either PNPase, SP1 or NFY subunit A (NFYA) is associated with a poor prognosis in liver cancer. In summary, our results show the relevance of SP1 and NFY in PNPT1 expression, and point to SP1/NFY and PNPase as possible targets in anti-cancer therapy.
Keywords: NFYA; PNPT1; SP1; liver cancer; mitochondria.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

