Tumor-Informed Approach Improved ctDNA Detection Rate in Resected Pancreatic Cancer
- PMID: 36232820
- PMCID: PMC9570468
- DOI: 10.3390/ijms231911521
Tumor-Informed Approach Improved ctDNA Detection Rate in Resected Pancreatic Cancer
Abstract
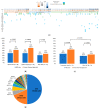
Pancreatic cancer is one of the cancers with very poor prognosis; there is an urgent need to identify novel biomarkers to improve its clinical outcomes. Circulating tumor DNA (ctDNA) from liquid biopsy has arisen as a promising biomarker for cancer detection and surveillance. However, it is known that the ctDNA detection rate in resected pancreatic cancer is low compared with other types of cancer. In this study, we collected paired tumor and plasma samples from 145 pancreatic cancer patients. Plasma samples were collected from 71 patients of treatment-naïve status and from 74 patients after neoadjuvant therapy (NAT). Genomic profiling of tumor DNA and plasma samples was conducted using targeted next-generation sequencing (NGS). Somatic mutations were detected in 85% (123/145) of tumors. ctDNA was detected in 39% (28/71) and 31% (23/74) of treatment-naïve and after-NAT groups, respectively, without referring to the information of tumor profiles. With a tumor-informed approach (TIA), ctDNA detection rate improved to 56% (40/71) and 36% (27/74) in treatment-naïve and after-NAT groups, respectively, with the detection rate significantly improved (p = 0.0165) among the treatment-naïve group compared to the after-NAT group. Cases who had detectable plasma ctDNA concordant to the corresponding tumor showed significantly shorter recurrence-free survival (RFS) (p = 0.0010). We demonstrated that TIA improves ctDNA detection rate in pancreatic cancer, and that ctDNA could be a potential prognostic biomarker for recurrence risk prediction.
Keywords: cancer prognosis; cell-free DNA; circulating tumor DNA; liquid biopsy; neoadjuvant therapy; next-generation sequencing; pancreatic cancer; tumor-informed approach.
Conflict of interest statement
Y.N. is a stockholder and a scientific advisor with OncoTherapy Science, Inc. S.-K.L. is a scientific advisor with Cancer Precision Medicine, Inc. No potential conflict of interest was disclosed by the other authors.
Figures




References
-
- Groot V.P., Rezaee N., Wu W., Cameron J.L., Fishman E.K., Hruban R.H., Weiss M.J., Zheng L., Wolfgang C.L., He J. Patterns, Timing, and Predictors of Recurrence Following Pancreatectomy for Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2018;267:936–945. doi: 10.1097/SLA.0000000000002234. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

