Rho/SRF Inhibitor Modulates Mitochondrial Functions
- PMID: 36232837
- PMCID: PMC9570101
- DOI: 10.3390/ijms231911536
Rho/SRF Inhibitor Modulates Mitochondrial Functions
Abstract
CCG-1423 is a Rho A pathway inhibitor that has been reported to inhibit Rho/SRF-mediated transcriptional regulation. Serum response factor and its cofactors, which include ternary complex factors and myocardin-related transcription factors, regulate various cellular functions. In this study, we observed that CCG-1423 modulates the mitochondrial functions. The effect of this small molecule drug was determined by measuring mitochondrial function using an XFe96 Analyzer and an Oxygraph 2k (O2k) high-resolution respirometer. CCG-1423 treatment significantly reduced oxidative phosphorylation in a dose-dependent manner. However, CCG-1423 increased the glycolytic rate. We also observed that histone 4 at lysine-16 underwent hyperacetylation with the treatment of this drug. Immunolabeling with F-actin and MitoTracker revealed the alteration in the actin cytoskeleton and mitochondria. Taken together, our findings highlight a critical role of CCG-1423 in inhibiting the transcription of SRF/p49 and PGC-1α, β, resulting in the downregulation of mitochondrial genes, leading to the repression of mitochondrial oxidative phosphorylation and overall ATP reduction. This study provides a better understanding of the effects of CCG-1423 on mitochondria, which may be useful for the assessment of the potential clinical application of CCG-1423 and its derivatives.
Keywords: CCG-1423; acetylation; mitochondrial function; serum response factor.
Conflict of interest statement
The authors declare no conflict of interest in regard to this manuscript.
Figures

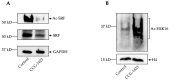
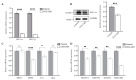
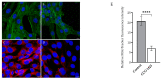
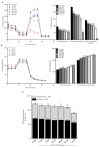


 = inhibition.
= inhibition.References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

