Oligonucleotide-Recognizing Topoisomerase Inhibitors (OTIs): Precision Gene Editors for Neurodegenerative Diseases?
- PMID: 36232843
- PMCID: PMC9570105
- DOI: 10.3390/ijms231911541
Oligonucleotide-Recognizing Topoisomerase Inhibitors (OTIs): Precision Gene Editors for Neurodegenerative Diseases?
Abstract
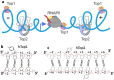
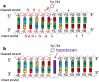
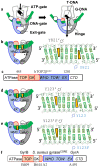
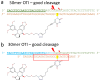
Topoisomerases are essential enzymes that recognize and modify the topology of DNA to allow DNA replication and transcription to take place. Topoisomerases are divided into type I topoisomerases, that cleave one DNA strand to modify DNA topology, and type II, that cleave both DNA strands. Topoisomerases normally rapidly religate cleaved-DNA once the topology has been modified. Topoisomerases do not recognize specific DNA sequences, but actively cleave positively supercoiled DNA ahead of transcription bubbles or replication forks, and negative supercoils (or precatenanes) behind, thus allowing the unwinding of the DNA-helix to proceed (during both transcription and replication). Drugs that stabilize DNA-cleavage complexes with topoisomerases produce cytotoxic DNA damage and kill fast-dividing cells; they are widely used in cancer chemotherapy. Oligonucleotide-recognizing topoisomerase inhibitors (OTIs) have given drugs that stabilize DNA-cleavage complexes specificity by linking them to either: (i) DNA duplex recognizing triplex forming oligonucleotide (TFO-OTIs) or DNA duplex recognizing pyrrole-imidazole-polyamides (PIP-OTIs) (ii) or by conventional Watson-Crick base pairing (WC-OTIs). This converts compounds from indiscriminate DNA-damaging drugs to highly specific targeted DNA-cleaving OTIs. Herein we propose simple strategies to enable DNA-duplex strand invasion of WC-OTIs giving strand-invading SI-OTIs. This will make SI-OTIs similar to the guide RNAs of CRISPR/Cas9 nuclease bacterial immune systems. However, an important difference between OTIs and CRISPR/Cas9, is that OTIs do not require the introduction of foreign proteins into cells. Recent successful oligonucleotide therapeutics for neurodegenerative diseases suggest that OTIs can be developed to be highly specific gene editing agents for DNA lesions that cause neurodegenerative diseases.
Keywords: CRISPR/Cas9; camptothecin; etoposide; gene editing; inhibitors; topoisomerases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

