Integration of QTL Mapping and Whole Genome Sequencing Identifies Candidate Genes for Alkalinity Tolerance in Rice (Oryza sativa)
- PMID: 36233092
- PMCID: PMC9569586
- DOI: 10.3390/ijms231911791
Integration of QTL Mapping and Whole Genome Sequencing Identifies Candidate Genes for Alkalinity Tolerance in Rice (Oryza sativa)
Abstract
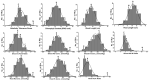
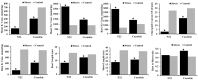
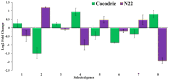
Soil alkalinity is an important stressor that impairs crop growth and development, resulting in reduced crop productivity. Unlike salinity stress, research efforts to understand the mechanism of plant adaptation to alkaline stress is limited in rice, a major staple food for the world population. We evaluated a population of 193 recombinant inbred lines (RIL) developed from a cross between Cocodrie and N22 under alkaline stress at the seedling stage. Using a linkage map consisting of 4849 SNP markers, 42 additive QTLs were identified. There were seven genomic regions where two or more QTLs for multiple traits colocalized. Three important QTL clusters were targeted, and several candidate genes were identified based on high impact variants using whole genome sequences (WGS) of both parents and differential expression in response to alkalinity stress. These genes included two expressed protein genes, the glucan endo-1,3-beta-glucosidase precursor, F-box domain-containing proteins, double-stranded RNA-binding motif-containing protein, aquaporin protein, receptor kinase-like protein, semialdehyde hydrogenase, and NAD-binding domain-containing protein genes. Tolerance to alkaline stress in Cocodrie was most likely due to the low Na+/K+ ratio resulting from reduced accumulation of Na+ ions and higher accumulation of K+ in roots and shoots. Our study demonstrated the utility of integrating QTL mapping with WGS to identify the candidate genes in the QTL regions. The QTLs and candidate genes originating from the tolerant parent Cocodrie should be targeted for introgression to improve alkalinity tolerance in rice and to elucidate the molecular basis of alkali tolerance.
Keywords: Na+/K+ ratio; Oryza sativa; abiotic stress; genotyping-by-sequencing; quantitative trait loci; seedling stage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Rao G.J.N., Reddy J.N., Variar M., Mahender A. Molecular breeding to improve plant resistance to abiotic stresses. In: Al-Khayri J.M., Jain S.M., Johnson D.V., editors. Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits. Springer International Publishing; Cham, Switzerland: 2016. pp. 283–326.
-
- Bray E.A., Bailey-Serres J., Weretilnyk E. Responses to abiotic stress. In: Gruissem W., Jones R., editors. Biochemistry & Molecular Biology of Plants. American Society of Plant Biologists; Rockville, MD, USA: 2000. pp. 1158–1249.
-
- Martinez-Beltran J., Manzur C.L. Overview of salinity problems in the world and FAO strategies to address the problem; Proceedings of the International Salinity Forum; Riverside, CA, USA. 25–27 April 2005; pp. 311–313.
-
- Qadir M., Quillerou E., Nangia V., Murtaza G., Singh M., Thomas R., Drechsel P., Noble A. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum. 2014;38:282–295. doi: 10.1111/1477-8947.12054. - DOI
-
- Chinnusamy V., Jagendorf A., Zhu J. Understanding and improving salt tolerance in plants. Crop Sci. 2020;45:437–448. doi: 10.2135/cropsci2005.0437. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

