Data Integration-Possibilities of Molecular and Clinical Data Fusion on the Example of Thyroid Cancer Diagnostics
- PMID: 36233181
- PMCID: PMC9569592
- DOI: 10.3390/ijms231911880
Data Integration-Possibilities of Molecular and Clinical Data Fusion on the Example of Thyroid Cancer Diagnostics
Abstract
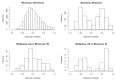
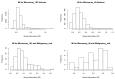
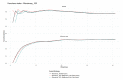
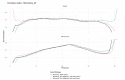
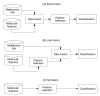
(1) Background: The data from independent gene expression sources may be integrated for the purpose of molecular diagnostics of cancer. So far, multiple approaches were described. Here, we investigated the impacts of different data fusion strategies on classification accuracy and feature selection stability, which allow the costs of diagnostic tests to be reduced. (2) Methods: We used molecular features (gene expression) combined with a feature extracted from the independent clinical data describing a patient's sample. We considered the dependencies between selected features in two data fusion strategies (early fusion and late fusion) compared to classification models based on molecular features only. We compared the best accuracy classification models in terms of the number of features, which is connected to the potential cost reduction of the diagnostic classifier. (3) Results: We show that for thyroid cancer, the extracted clinical feature is correlated with (but not redundant to) the molecular data. The usage of data fusion allows a model to be obtained with similar or even higher classification quality (with a statistically significant accuracy improvement, a p-value below 0.05) and with a reduction in molecular dimensionality of the feature space from 15 to 3-8 (depending on the feature selection method). (4) Conclusions: Both strategies give comparable quality results, but the early fusion method provides better feature selection stability.
Keywords: bioinformatics; biomarkers; cancer; classification; data fusion; data integration; thyroid cancer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Hira Z.M., Gillies D.F. A Review of Feature Selection and Feature Extraction Methods Applied on Microarray Data. [(accessed on 5 April 2020)]. Available online: https://www.hindawi.com/journals/abi/2015/198363/ - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

