Analysis of Protein-Protein Interactions for Intermolecular Bond Prediction
- PMID: 36234723
- PMCID: PMC9572624
- DOI: 10.3390/molecules27196178
Analysis of Protein-Protein Interactions for Intermolecular Bond Prediction
Abstract
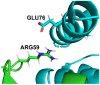
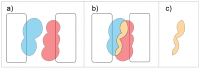
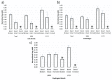
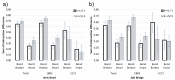
Protein-protein interactions often involve a complex system of intermolecular interactions between residues and atoms at the binding site. A comprehensive exploration of these interactions can help reveal key residues involved in protein-protein recognition that are not obvious using other protein analysis techniques. This paper presents and extends DiffBond, a novel method for identifying and classifying intermolecular bonds while applying standard definitions of bonds in chemical literature to explain protein interactions. DiffBond predicted intermolecular bonds from four protein complexes: Barnase-Barstar, Rap1a-raf, SMAD2-SMAD4, and a subset of complexes formed from three-finger toxins and nAChRs. Based on validation through manual literature search and through comparison of two protein complexes from the SKEMPI dataset, DiffBond was able to identify intermolecular ionic bonds and hydrogen bonds with high precision and recall, and identify salt bridges with high precision. DiffBond predictions on bond existence were also strongly correlated with observations of Gibbs free energy change and electrostatic complementarity in mutational experiments. DiffBond can be a powerful tool for predicting and characterizing influential residues in protein-protein interactions, and its predictions can support research in mutational experiments and drug design.
Keywords: DiffBond; bond classifier; intermolecular bond prediction; ionic bond identificatio.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

