Applications of MALDI-MS/MS-Based Proteomics in Biomedical Research
- PMID: 36234736
- PMCID: PMC9570737
- DOI: 10.3390/molecules27196196
Applications of MALDI-MS/MS-Based Proteomics in Biomedical Research
Abstract
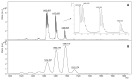
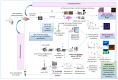
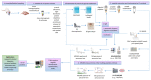
Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) is one of the most widely used techniques in proteomics to achieve structural identification and characterization of proteins and peptides, including their variety of proteoforms due to post-translational modifications (PTMs) or protein-protein interactions (PPIs). MALDI-MS and MALDI tandem mass spectrometry (MS/MS) have been developed as analytical techniques to study small and large molecules, offering picomole to femtomole sensitivity and enabling the direct analysis of biological samples, such as biofluids, solid tissues, tissue/cell homogenates, and cell culture lysates, with a minimized procedure of sample preparation. In the last decades, structural identification of peptides and proteins achieved by MALDI-MS/MS helped researchers and clinicians to decipher molecular function, biological process, cellular component, and related pathways of the gene products as well as their involvement in pathogenesis of diseases. In this review, we highlight the applications of MALDI ionization source and tandem approaches for MS for analyzing biomedical relevant peptides and proteins. Furthermore, one of the most relevant applications of MALDI-MS/MS is to provide "molecular pictures", which offer in situ information about molecular weight proteins without labeling of potential targets. Histology-directed MALDI-mass spectrometry imaging (MSI) uses MALDI-ToF/ToF or other MALDI tandem mass spectrometers for accurate sequence analysis of peptide biomarkers and biological active compounds directly in tissues, to assure complementary and essential spatial data compared with those obtained by LC-ESI-MS/MS technique.
Keywords: MALDI; biomarkers; biomedical research; proteomics; tandem mass spectrometry (MS/MS).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
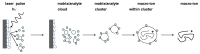


References
-
- Karas M., Bachmann D., Bahr U., Hillenkamp F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int. J. Mass Spectrom. Ion Processes. 1987;78:53–68. doi: 10.1016/0168-1176(87)87041-6. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

