Small Molecules for Enhancing the Precision and Safety of Genome Editing
- PMID: 36234804
- PMCID: PMC9573751
- DOI: 10.3390/molecules27196266
Small Molecules for Enhancing the Precision and Safety of Genome Editing
Abstract
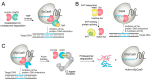
Clustered regularly interspaced short palindromic repeats (CRISPR)-based genome-editing technologies have revolutionized biology, biotechnology, and medicine, and have spurred the development of new therapeutic modalities. However, there remain several barriers to the safe use of CRISPR technologies, such as unintended off-target DNA cleavages. Small molecules are important resources to solve these problems, given their facile delivery and fast action to enable temporal control of the CRISPR systems. Here, we provide a comprehensive overview of small molecules that can precisely modulate CRISPR-associated (Cas) nucleases and guide RNAs (gRNAs). We also discuss the small-molecule control of emerging genome editors (e.g., base editors) and anti-CRISPR proteins. These molecules could be used for the precise investigation of biological systems and the development of safer therapeutic modalities.
Keywords: CRISPR; Cas nuclease; genome editing; guide RNA; small molecule; specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Targeted genome editing in human cells using CRISPR/Cas nucleases and truncated guide RNAs.Methods Enzymol. 2014;546:21-45. doi: 10.1016/B978-0-12-801185-0.00002-7. Methods Enzymol. 2014. PMID: 25398334
-
Conditional Control of CRISPR/Cas9 Function.Angew Chem Int Ed Engl. 2016 Apr 25;55(18):5394-9. doi: 10.1002/anie.201511441. Epub 2016 Mar 21. Angew Chem Int Ed Engl. 2016. PMID: 26996256 Review.
-
[CRISPR/CAS9, the King of Genome Editing Tools].Mol Biol (Mosk). 2017 Jul-Aug;51(4):582-594. doi: 10.7868/S0026898417040036. Mol Biol (Mosk). 2017. PMID: 28900076 Review. Russian.
-
Strategies for Optimization of the Clustered Regularly Interspaced Short Palindromic Repeat-Based Genome Editing System for Enhanced Editing Specificity.Hum Gene Ther. 2022 Apr;33(7-8):358-370. doi: 10.1089/hum.2021.283. Epub 2022 Mar 1. Hum Gene Ther. 2022. PMID: 34963339 Review.
-
Evaluating and Enhancing Target Specificity of Gene-Editing Nucleases and Deaminases.Annu Rev Biochem. 2019 Jun 20;88:191-220. doi: 10.1146/annurev-biochem-013118-111730. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883196 Review.
Cited by
-
Optical Genome Mapping Reveals Genomic Alterations upon Gene Editing in hiPSCs: Implications for Neural Tissue Differentiation and Brain Organoid Research.Cells. 2024 Mar 14;13(6):507. doi: 10.3390/cells13060507. Cells. 2024. PMID: 38534351 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

