Genome-Wide Identification and Expression Analysis of the Zinc Finger Protein Gene Subfamilies under Drought Stress in Triticum aestivum
- PMID: 36235376
- PMCID: PMC9572532
- DOI: 10.3390/plants11192511
Genome-Wide Identification and Expression Analysis of the Zinc Finger Protein Gene Subfamilies under Drought Stress in Triticum aestivum
Abstract
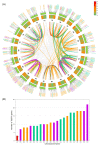
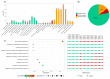
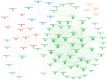
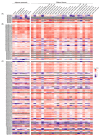
The zinc finger protein (ZFP) family is one of plants' most diverse family of transcription factors. These proteins with finger-like structural domains have been shown to play a critical role in plant responses to abiotic stresses such as drought. This study aimed to systematically characterize Triticum aestivum ZFPs (TaZFPs) and understand their roles under drought stress. A total of 9 TaC2H2, 38 TaC3HC4, 79 TaCCCH, and 143 TaPHD were identified, which were divided into 4, 7, 12, and 14 distinct subgroups based on their phylogenetic relationships, respectively. Segmental duplication dominated the evolution of four subfamilies and made important contributions to the large-scale amplification of gene families. Syntenic relationships, gene duplications, and Ka/Ks result consistently indicate a potential strong purifying selection on TaZFPs. Additionally, TaZFPs have various abiotic stress-associated cis-acting regulatory elements and have tissue-specific expression patterns showing different responses to drought and heat stress. Therefore, these genes may play multiple functions in plant growth and stress resistance responses. This is the first comprehensive genome-wide analysis of ZFP gene families in T. aestivum to elucidate the basis of their function and resistance mechanisms, providing a reference for precise manipulation of genetic engineering for drought resistance in T. aestivum.
Keywords: Triticum aestivum; abiotic stresses; drought stress; expression pattern; genome-wide identification; zinc finger proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Rehaman A., Mishra A.K., Ferdose A., Per T.S., Hanief M., Jan A.T., Asgher M. Melatonin in Plant Defense against Abiotic Stress. Forests. 2021;12:1404. doi: 10.3390/f12101404. - DOI
-
- Naeem M., Shahzad K., Saqib S., Shahzad A., Nasrullah, Younas M., Afridi M.I. The Solanum Melongena COP1LIKE Manipulates Fruit Ripening and Flowering Time in Tomato (Solanum lycopersicum) Plant Growth Regul. 2022;96:369–382. doi: 10.1007/s10725-021-00785-7. - DOI
LinkOut - more resources
Full Text Sources

