The C-Terminal Region of SLIM1 Transcription Factor Is Required for Sulfur Deficiency Response
- PMID: 36235462
- PMCID: PMC9573389
- DOI: 10.3390/plants11192595
The C-Terminal Region of SLIM1 Transcription Factor Is Required for Sulfur Deficiency Response
Abstract
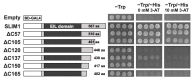
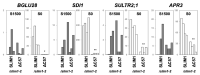
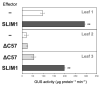
Sulfur LIMitation1 (SLIM1) transcription factor coordinates gene expression in plants in response to sulfur deficiency (-S). SLIM1 belongs to the family of plant-specific EIL transcription factors with EIN3 and EIL1, which regulate the ethylene-responsive gene expression. The EIL domains consist of DNA binding and dimerization domains highly conserved among EIL family members, while the N- and C-terminal regions are structurally variable and postulated to have regulatory roles in this protein family, such that the EIN3 C-terminal region is essential for its ethylene-responsive activation. In this study, we focused on the roles of the SLIM1 C-terminal region. We examined the transactivation activity of the full-length and the truncated SLIM1 in yeast and Arabidopsis. The full-length SLIM1 and the truncated form of SLIM1 with a deletion of C-terminal 106 amino acids (ΔC105) transactivated the reporter gene expression in yeast when they were fused to the GAL4 DNA binding domain, whereas the deletion of additional 15 amino acids to remove the C-terminal 120 amino acids (ΔC120) eliminated such an activity, identifying the necessity of that 15-amino-acid segment for transactivation. In the Arabidopsis slim1-2 mutant, the transcript levels of SULTR1;2 sulfate transporter and the GFP expression derived from the SULTR1;2 promoter-GFP (PSULTR1;2-GFP) transgene construct were restored under -S by introducing the full-length SLIM1, but not with the C-terminal truncated forms ΔC105 and ΔC57. Furthermore, the transcript levels of -S-responsive genes were restored concomitantly with an increase in glutathione accumulation in the complementing lines with the full-length SLIM1 but not with ΔC57. The C-terminal 57 amino acids of SLIM1 were also shown to be necessary for transactivation of a -S-inducible gene, SHM7/MSA1, in a transient expression system using the SHM7/MSA1 promoter-GUS as a reporter. These findings suggest that the C-terminal region is essential for the SLIM1 activity.
Keywords: Arabidopsis thaliana; SLIM1 transcription factor; sulfate assimilation; sulfate transporter; sulfur deficiency.
Conflict of interest statement
The authors have no conflict of interest to declare. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Long S.R., Kahn M., Seefeldt L., Tsay Y.F., Kopriva S. Nitrogen and Sulfur. In: Buchana B.B., Gruissem W., Jones R.L., editors. Biochemistry & Molecular Biology of Plants. 2nd ed. Wiley Blackwell; Oxford, UK: 2015. pp. 746–768.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

