Knot Factories with Helical Geometry Enhance Knotting and Induce Handedness to Knots
- PMID: 36236148
- PMCID: PMC9572405
- DOI: 10.3390/polym14194201
Knot Factories with Helical Geometry Enhance Knotting and Induce Handedness to Knots
Abstract
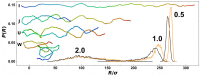
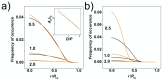
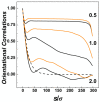
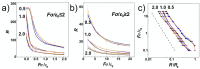
We performed molecular dynamics simulations of DNA polymer chains confined in helical nano-channels under compression in order to explore the potential of knot-factories with helical geometry to produce knots with a preferred handedness. In our simulations, we explore mutual effect of the confinement strength and compressive forces in a range covering weak, intermediate and strong confinement together with weak and strong compressive forces. The results find that while the common metrics of polymer chain in cylindrical and helical channels are very similar, the DNA in helical channels exhibits greatly different topology in terms of chain knottedness, writhe and handedness of knots. The results show that knots with a preferred chirality in terms of average writhe can be produced by using channels with a chosen handedness.
Keywords: DNA; chirality; compression; helical; knot factory; knots; molecular dynamics; nano-channels; polymer; topology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Knot Formation on DNA Pushed Inside Chiral Nanochannels.Polymers (Basel). 2023 Oct 22;15(20):4185. doi: 10.3390/polym15204185. Polymers (Basel). 2023. PMID: 37896430 Free PMC article.
-
Channels with Helical Modulation Display Stereospecific Sensitivity for Chiral Superstructures.Polymers (Basel). 2021 Oct 28;13(21):3726. doi: 10.3390/polym13213726. Polymers (Basel). 2021. PMID: 34771282 Free PMC article.
-
Interplay between writhe and knotting for swollen and compact polymers.J Chem Phys. 2009 Oct 21;131(15):154902. doi: 10.1063/1.3244643. J Chem Phys. 2009. PMID: 20568879
-
New biologically motivated knot table.Biochem Soc Trans. 2013 Apr;41(2):606-11. doi: 10.1042/BST20120278. Biochem Soc Trans. 2013. PMID: 23514162 Review.
-
Molecular Knots.Angew Chem Int Ed Engl. 2017 Sep 4;56(37):11166-11194. doi: 10.1002/anie.201702531. Epub 2017 Aug 16. Angew Chem Int Ed Engl. 2017. PMID: 28477423 Free PMC article. Review.
Cited by
-
Knot Formation on DNA Pushed Inside Chiral Nanochannels.Polymers (Basel). 2023 Oct 22;15(20):4185. doi: 10.3390/polym15204185. Polymers (Basel). 2023. PMID: 37896430 Free PMC article.
References
-
- Amin S. Knot Formation on Nanochannel-Confined DNA and Flow-Assisted DNA Disentanglement and Linearization. McGill University; Montreal, QC, Canada: 2018.
-
- Khorshid A., Amin S., Zhang Z., Sakaue T., Reisner W.W. Nonequilibrium Dynamics of Nanochannel Confined DNA. Macromolecules. 2016;49:1933–1940. doi: 10.1021/acs.macromol.5b02240. - DOI
-
- Ma Z., Dorfman K.D. Diffusion of Knots along DNA Confined in Nanochannels. Macromolecules. 2020;53:6461–6468. doi: 10.1021/acs.macromol.0c00561. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

