Atrophic skeletal muscle fibre-derived small extracellular vesicle miR-690 inhibits satellite cell differentiation during ageing
- PMID: 36237168
- PMCID: PMC9745557
- DOI: 10.1002/jcsm.13106
Atrophic skeletal muscle fibre-derived small extracellular vesicle miR-690 inhibits satellite cell differentiation during ageing
Abstract
Background: Sarcopenia is a common and progressive skeletal muscle disorder characterized by atrophic muscle fibres and contractile dysfunction. Accumulating evidence shows that the number and function of satellite cells (SCs) decline and become impaired during ageing, which may contribute to impaired regenerative capacity. A series of myokines/small extracellular vesicles (sEVs) released from muscle fibres regulate metabolism in muscle and extramuscular tissues in an autocrine/paracrine/endocrine manner during muscle atrophy. It is still unclear whether myokines/sEVs derived from muscle fibres can affect satellite cell function during ageing.
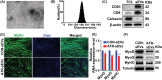
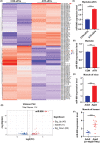
Methods: Aged mice were used to investigate changes in the myogenic capacity of SCs during ageing-induced muscle atrophy. The effects of atrophic myotube-derived sEVs on satellite cell differentiation were investigated by biochemical methods and immunofluorescence staining. Small RNA sequencing was performed to identify differentially expressed sEV microRNAs (miRNAs) between the control myotubes and atrophic myotubes. The target genes of the miRNA were predicted by bioinformatics analysis and verified by luciferase activity assays. The effects of identified miRNA on the myogenic capacity of SCs in vivo were investigated by intramuscular injection of adeno-associated virus (AAV) to overexpress or silence miRNA in skeletal muscle.
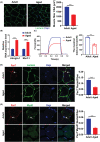
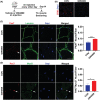
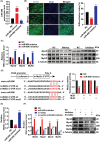
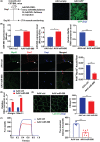
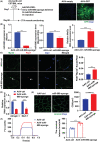
Results: Our study showed that the myogenic capacity of SCs was significantly decreased (50%, n = 6, P < 0.001) in the tibialis anterior muscle of aged mice. We showed that atrophic myotube-derived sEVs inhibited satellite cell differentiation in vitro (n = 3, P < 0.001) and in vivo (35%, n = 6, P < 0.05). We also found that miR-690 was the most highly enriched miRNA among all the screened sEV miRNAs in atrophic myotubes [Log2 (Fold Change) = 7, P < 0.001], which was verified in the atrophic muscle of aged mice (threefold, n = 6, P < 0.001) and aged men with mean age of 71 ± 5.27 years (2.8-fold, n = 10, P < 0.001). MiR-690 can inhibit myogenic capacity of SCs by targeting myocyte enhancer factor 2, including Mef2a, Mef2c and Mef2d, in vitro (n = 3, P < 0.05) and in vivo (n = 6, P < 0.05). Specific silencing of miR-690 in the muscle can promote satellite cell differentiation (n = 6, P < 0.001) and alleviate muscle atrophy in aged mice (n = 6, P < 0.001).
Conclusions: Our study demonstrated that atrophic muscle fibre-derived sEV miR-690 may inhibit satellite cell differentiation by targeting myocyte enhancer factor 2 during ageing.
Keywords: Muscle fibre; Myogenic capacity; Sarcopenia; Satellite cells; Small extracellular vesicle; miR-690.
© 2022 The Authors. Journal of Cachexia, Sarcopenia and Muscle published by John Wiley & Sons Ltd on behalf of Society on Sarcopenia, Cachexia and Wasting Disorders.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures

References
-
- Mayhew AJ, Amog K, Phillips S, Parise G, McNicholas PD, de Souza RJ, Thabane L, Raina P. The prevalence of sarcopenia in community‐dwelling older adults, an exploration of differences between studies and within definitions: A systematic review and meta‐analyses. Age Ageing 2019;48:48–56. - PubMed
-
- Cruz‐Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyère O, Cederholm T, Cooper C, Landi F, Rolland Y, Sayer AA, Schneider SM, Sieber CC, Topinkova E, Vandewoude M, Visser M, Zamboni M, Writing Group for the European Working Group on Sarcopenia in Older People 2 (EWGSOP2), and the Extended Group for EWGSOP2 , Bautmans I, Baeyens JP, Cesari M, Cherubini A, Kanis J, Maggio M, Martin F, Michel JP, Pitkala K, Reginster JY, Rizzoli R, Sánchez‐Rodríguez D, Schols J. Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 2019;48:16–31. - PMC - PubMed
-
- Siriett V, Salerno MS, Berry C, Nicholas G, Bower R, Kambadur R, Sharma M. Antagonism of myostatin enhances muscle regeneration during sarcopenia. Mole Ther:J Am Soc Gene Ther 2007;15:1463–1470. - PubMed
-
- Lees SJ, Rathbone CR, Booth FW. Age‐associated decrease in muscle precursor cell differentiation. Am J Physiol Cell Physiol 2006;290:C609–C615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

