Discovery of New Zika Protease and Polymerase Inhibitors through the Open Science Collaboration Project OpenZika
- PMID: 36239304
- PMCID: PMC9923514
- DOI: 10.1021/acs.jcim.2c00596
Discovery of New Zika Protease and Polymerase Inhibitors through the Open Science Collaboration Project OpenZika
Abstract
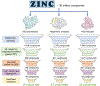
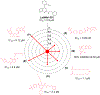
The Zika virus (ZIKV) is a neurotropic arbovirus considered a global threat to public health. Although there have been several efforts in drug discovery projects for ZIKV in recent years, there are still no antiviral drugs approved to date. Here, we describe the results of a global collaborative crowdsourced open science project, the OpenZika project, from IBM's World Community Grid (WCG), which integrates different computational and experimental strategies for advancing a drug candidate for ZIKV. Initially, molecular docking protocols were developed to identify potential inhibitors of ZIKV NS5 RNA-dependent RNA polymerase (NS5 RdRp), NS3 protease (NS2B-NS3pro), and NS3 helicase (NS3hel). Then, a machine learning (ML) model was built to distinguish active vs inactive compounds for the cytoprotective effect against ZIKV infection. We performed three independent target-based virtual screening campaigns (NS5 RdRp, NS2B-NS3pro, and NS3hel), followed by predictions by the ML model and other filters, and prioritized a total of 61 compounds for further testing in enzymatic and phenotypic assays. This yielded five non-nucleoside compounds which showed inhibitory activity against ZIKV NS5 RdRp in enzymatic assays (IC50 range from 0.61 to 17 μM). Two compounds thermally destabilized NS3hel and showed binding affinity in the micromolar range (Kd range from 9 to 35 μM). Moreover, the compounds LabMol-301 inhibited both NS5 RdRp and NS2B-NS3pro (IC50 of 0.8 and 7.4 μM, respectively) and LabMol-212 thermally destabilized the ZIKV NS3hel (Kd of 35 μM). Both also protected cells from death induced by ZIKV infection in in vitro cell-based assays. However, while eight compounds (including LabMol-301 and LabMol-212) showed a cytoprotective effect and prevented ZIKV-induced cell death, agreeing with our ML model for prediction of this cytoprotective effect, no compound showed a direct antiviral effect against ZIKV. Thus, the new scaffolds discovered here are promising hits for future structural optimization and for advancing the discovery of further drug candidates for ZIKV. Furthermore, this work has demonstrated the importance of the integration of computational and experimental approaches, as well as the potential of large-scale collaborative networks to advance drug discovery projects for neglected diseases and emerging viruses, despite the lack of available direct antiviral activity and cytoprotective effect data, that reflects on the assertiveness of the computational predictions. The importance of these efforts rests with the need to be prepared for future viral epidemic and pandemic outbreaks.
Conflict of interest statement
Declaration of Interest
S.E. is founder and owner of Collaborations Pharmaceuticals, A.C.P. and F.U. is employee of Collaborations Pharmaceuticals, Inc. The remaining authors declare that there are no conflicts of interest.
Figures
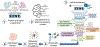
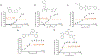



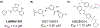
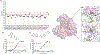
Similar articles
-
Identification of Zika virus NS2B-NS3 protease and NS5 polymerase inhibitors by structure-based virtual screening of FDA-approved drugs.J Biomol Struct Dyn. 2024 Sep;42(15):8073-8088. doi: 10.1080/07391102.2023.2242963. Epub 2023 Aug 1. J Biomol Struct Dyn. 2024. PMID: 37528667
-
Discovery of Bispecific Lead Compounds from Azadirachta indica against ZIKA NS2B-NS3 Protease and NS5 RNA Dependent RNA Polymerase Using Molecular Simulations.Molecules. 2022 Apr 15;27(8):2562. doi: 10.3390/molecules27082562. Molecules. 2022. PMID: 35458761 Free PMC article.
-
Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.J Virol. 2020 Oct 14;94(21):e00794-20. doi: 10.1128/JVI.00794-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796069 Free PMC article.
-
NS2B-NS3 protease inhibitors as promising compounds in the development of antivirals against Zika virus: A systematic review.J Med Virol. 2022 Feb;94(2):442-453. doi: 10.1002/jmv.27386. Epub 2021 Oct 20. J Med Virol. 2022. PMID: 34636434
-
The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection.Int J Mol Sci. 2024 Apr 16;25(8):4376. doi: 10.3390/ijms25084376. Int J Mol Sci. 2024. PMID: 38673962 Free PMC article. Review.
Cited by
-
Natural Compounds as Non-Nucleoside Inhibitors of Zika Virus Polymerase through Integration of In Silico and In Vitro Approaches.Pharmaceuticals (Basel). 2022 Nov 30;15(12):1493. doi: 10.3390/ph15121493. Pharmaceuticals (Basel). 2022. PMID: 36558945 Free PMC article.
-
Deciphering Structural Dynamics of Atherosclerosis Proteins: Insights from Crataegus oxyacantha Phytochemicals that Interceded Functional and Structural Changes in Targeted Atherosclerotic Proteins.ACS Omega. 2024 Nov 23;9(49):48159-48172. doi: 10.1021/acsomega.4c04975. eCollection 2024 Dec 10. ACS Omega. 2024. PMID: 39676950 Free PMC article.
-
Introducing CRAFT: The Center for Research and Advancement in Fragments and molecular Targets.ACS Med Chem Lett. 2024 Jul 23;15(8):1174-1177. doi: 10.1021/acsmedchemlett.4c00296. eCollection 2024 Aug 8. ACS Med Chem Lett. 2024. PMID: 39140068 Free PMC article.
-
The protein disulfide isomerase inhibitor 3-methyltoxoflavin inhibits Chikungunya virus.Bioorg Med Chem. 2023 Apr 1;83:117239. doi: 10.1016/j.bmc.2023.117239. Epub 2023 Mar 15. Bioorg Med Chem. 2023. PMID: 36940609 Free PMC article.
-
Evaluating Known Zika Virus NS2B-NS3 Protease Inhibitor Scaffolds via In Silico Screening and Biochemical Assays.Pharmaceuticals (Basel). 2023 Sep 19;16(9):1319. doi: 10.3390/ph16091319. Pharmaceuticals (Basel). 2023. PMID: 37765127 Free PMC article.
References
-
- Besnard M; Lastère S; Teissier A; Cao-Lormeau VM; Musso D Evidence of Perinatal Transmission of Zika Virus. Euro Surveill 2013, 19. - PubMed
-
- Musso D; Nhan T; Robin E; Roche C; Bierlaire D; Zisou K; Shan Yan A; Cao-Lormeau V; Broult J Potential for Zika Virus Transmission through Blood Transfusion Demonstrated during an Outbreak in French Polynesia, November 2013 to February 2014. Eurosurveillance 2014, 19, 20761. 10.2807/1560-7917.ES2014.19.14.20761. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

