CXCL12 defines lung endothelial heterogeneity and promotes distal vascular growth
- PMID: 36239312
- PMCID: PMC9687018
- DOI: 10.1242/dev.200909
CXCL12 defines lung endothelial heterogeneity and promotes distal vascular growth
Abstract
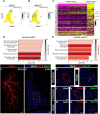
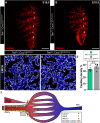
There is a growing amount of data uncovering the cellular diversity of the pulmonary circulation and mechanisms governing vascular repair after injury. However, the molecular and cellular mechanisms contributing to the morphogenesis and growth of the pulmonary vasculature during embryonic development are less clear. Importantly, deficits in vascular development lead to significant pediatric lung diseases, indicating a need to uncover fetal programs promoting vascular growth. To address this, we used a transgenic mouse reporter for expression of Cxcl12, an arterial endothelial hallmark gene, and performed single-cell RNA sequencing on isolated Cxcl12-DsRed+ endothelium to assess cellular heterogeneity within pulmonary endothelium. Combining cell annotation with gene ontology and histological analysis allowed us to segregate the developing artery endothelium into functionally and spatially distinct subpopulations. Expression of Cxcl12 is highest in the distal arterial endothelial subpopulation, a compartment enriched in genes for vascular development. Accordingly, disruption of CXCL12 signaling led to, not only abnormal branching, but also distal vascular hypoplasia. These data provide evidence for arterial endothelial functional heterogeneity and reveal conserved signaling mechanisms essential for pulmonary vascular development.
Keywords: Endothelium; Lung; Mouse; Single-cell RNA sequencing.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests L.R.Y. declares grants paid to her institution from the National Institutes of Health and the Orphan Disease Center at the University of Pennsylvania; royalties from UpToDate; consultancy fees and fees for participation on the Steering Committee of the InPedILD trial from Boehringer Ingelheim; and consultancy fees from Roche and Sanofi.
Figures





References
-
- Asosingh, K., Comhair, S., Mavrakis, L., Xu, W., Horton, D., Taylor, I., Tkachenko, S., Hu, B. and Erzurum, S. (2021). Single-cell transcriptomic profile of human pulmonary artery endothelial cells in health and pulmonary arterial hypertension. Sci. Rep. 11, 14714. 10.1038/s41598-021-94163-y - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

