The GARP complex prevents sterol accumulation at the trans-Golgi network during dendrite remodeling
- PMID: 36239632
- PMCID: PMC9577387
- DOI: 10.1083/jcb.202112108
The GARP complex prevents sterol accumulation at the trans-Golgi network during dendrite remodeling
Abstract
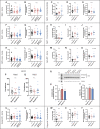
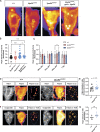
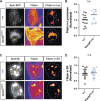
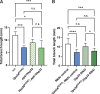
Membrane trafficking is essential for sculpting neuronal morphology. The GARP and EARP complexes are conserved tethers that regulate vesicle trafficking in the secretory and endolysosomal pathways, respectively. Both complexes contain the Vps51, Vps52, and Vps53 proteins, and a complex-specific protein: Vps54 in GARP and Vps50 in EARP. In Drosophila, we find that both complexes are required for dendrite morphogenesis during developmental remodeling of multidendritic class IV da (c4da) neurons. Having found that sterol accumulates at the trans-Golgi network (TGN) in Vps54KO/KO neurons, we investigated genes that regulate sterols and related lipids at the TGN. Overexpression of oxysterol binding protein (Osbp) or knockdown of the PI4K four wheel drive (fwd) exacerbates the Vps54KO/KO phenotype, whereas eliminating one allele of Osbp rescues it, suggesting that excess sterol accumulation at the TGN is, in part, responsible for inhibiting dendrite regrowth. These findings distinguish the GARP and EARP complexes in neurodevelopment and implicate vesicle trafficking and lipid transfer pathways in dendrite morphogenesis.
© 2022 O'Brien et al.
Figures
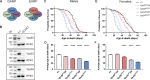
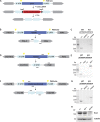


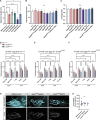

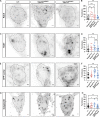

Comment in
-
Dendrite remodeling according to GARP.J Cell Biol. 2023 Jan 2;222(1):e202211072. doi: 10.1083/jcb.202211072. Epub 2022 Dec 22. J Cell Biol. 2023. PMID: 36547519 Free PMC article.
References
-
- Castrillon, D.H., Gonczy P., Alexander S., Rawson R., Eberhart C.G., Viswanathan S., DiNardo S., and Wasserman S.A.. 1993. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: Characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 135:489–505. 10.1093/genetics/135.2.489 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

