Nascent polypeptide-Associated Complex and Signal Recognition Particle have cardiac-specific roles in heart development and remodeling
- PMID: 36240221
- PMCID: PMC9604979
- DOI: 10.1371/journal.pgen.1010448
Nascent polypeptide-Associated Complex and Signal Recognition Particle have cardiac-specific roles in heart development and remodeling
Abstract
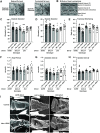
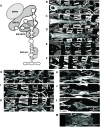
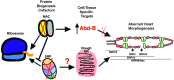
Establishing a catalog of Congenital Heart Disease (CHD) genes and identifying functional networks would improve our understanding of its oligogenic underpinnings. Our studies identified protein biogenesis cofactors Nascent polypeptide-Associated Complex (NAC) and Signal-Recognition-Particle (SRP) as disease candidates and novel regulators of cardiac differentiation and morphogenesis. Knockdown (KD) of the alpha- (Nacα) or beta-subunit (bicaudal, bic) of NAC in the developing Drosophila heart disrupted cardiac developmental remodeling resulting in a fly with no heart. Heart loss was rescued by combined KD of Nacα with the posterior patterning Hox gene Abd-B. Consistent with a central role for this interaction in cardiogenesis, KD of Nacα in cardiac progenitors derived from human iPSCs impaired cardiac differentiation while co-KD with human HOXC12 and HOXD12 rescued this phenotype. Our data suggest that Nacα KD preprograms cardioblasts in the embryo for abortive remodeling later during metamorphosis, as Nacα KD during translation-intensive larval growth or pupal remodeling only causes moderate heart defects. KD of SRP subunits in the developing fly heart produced phenotypes that targeted specific segments and cell types, again suggesting cardiac-specific and spatially regulated activities. Together, we demonstrated directed function for NAC and SRP in heart development, and that regulation of NAC function depends on Hox genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Lage K, Greenway SC, Rosenfeld JA, Wakimoto H, Gorham JM, Segre AV, et al.. Genetic and environmental risk factors in congenital heart disease functionally converge in protein networks driving heart development. Proc Natl Acad Sci U S A. 2012;109(35):14035–40. doi: 10.1073/pnas.1210730109 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

