Ensemble-function relationships to dissect mechanisms of enzyme catalysis
- PMID: 36240280
- PMCID: PMC9565801
- DOI: 10.1126/sciadv.abn7738
Ensemble-function relationships to dissect mechanisms of enzyme catalysis
Abstract
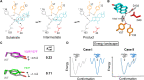
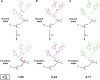
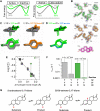
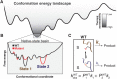
Decades of structure-function studies have established our current extensive understanding of enzymes. However, traditional structural models are snapshots of broader conformational ensembles of interchanging states. We demonstrate the need for conformational ensembles to understand function, using the enzyme ketosteroid isomerase (KSI) as an example. Comparison of prior KSI cryogenic x-ray structures suggested deleterious mutational effects from a misaligned oxyanion hole catalytic residue. However, ensemble information from room-temperature x-ray crystallography, combined with functional studies, excluded this model. Ensemble-function analyses can deconvolute effects from altering the probability of occupying a state (P-effects) and changing the reactivity of each state (k-effects); our ensemble-function analyses revealed functional effects arising from weakened oxyanion hole hydrogen bonding and substrate repositioning within the active site. Ensemble-function studies will have an integral role in understanding enzymes and in meeting the future goals of a predictive understanding of enzyme catalysis and engineering new enzymes.
Figures

References
-
- J. M. Berg, J. L. Tymoczko, L. Stryer, J. M. Berg, J. L. Tymoczko, L. Stryer, Biochemistry (W H Freeman, ed. 5, 2002).
-
- A. Fersht, Enzyme Structure and Mechanism (W.H. Freeman, 1985).
-
- Kim D.-H., Jang D. S., Nam G. H., Choi G., Kim J.-S., Ha N.-C., Kim M.-S., Oh B.-H., Choi K. Y., Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B. Biochemistry 39, 4581–4589 (2000). - PubMed
-
- Lawson S. L., Wakarchuk W. W., Withers S. G., Effects of both shortening and lengthening the active site nucleophile of bacillus circulans xylanase on catalytic activity. Biochemistry 35, 10110–10118 (1996). - PubMed

