Integrated flow cytometry and sequencing to reconstruct evolutionary patterns from dysplasia to acute myeloid leukemia
- PMID: 36240453
- PMCID: PMC9811200
- DOI: 10.1182/bloodadvances.2022008141
Integrated flow cytometry and sequencing to reconstruct evolutionary patterns from dysplasia to acute myeloid leukemia
Abstract
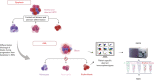
Clonal evolution in acute myeloid leukemia (AML) originates long before diagnosis and is a dynamic process that may affect survival. However, it remains uninvestigated during routine diagnostic workups. We hypothesized that the mutational status of bone marrow dysplastic cells and leukemic blasts, analyzed at the onset of AML using integrated multidimensional flow cytometry (MFC) immunophenotyping and fluorescence-activated cell sorting (FACS) with next-generation sequencing (NGS), could reconstruct leukemogenesis. Dysplastic cells were detected by MFC in 285 of 348 (82%) newly diagnosed patients with AML. Presence of dysplasia according to MFC and World Health Organization criteria had no prognostic value in older adults. NGS of dysplastic cells and blasts isolated at diagnosis identified 3 evolutionary patterns: stable (n = 12 of 21), branching (n = 4 of 21), and clonal evolution (n = 5 of 21). In patients achieving complete response (CR), integrated MFC and FACS with NGS showed persistent measurable residual disease (MRD) in phenotypically normal cell types, as well as the acquisition of genetic traits associated with treatment resistance. Furthermore, whole-exome sequencing of dysplastic and leukemic cells at diagnosis and of MRD uncovered different clonal involvement in dysplastic myelo-erythropoiesis, leukemic transformation, and chemoresistance. Altogether, we showed that it is possible to reconstruct leukemogenesis in ∼80% of patients with newly diagnosed AML, using techniques other than single-cell multiomics.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: F.P. received honoraria and research funding from Oryzon, Janssen, Bristol-Myers Squibb (BMS)/Celgene. R.A. serves as a member of the board of directors advisory committees for Incyte Corporation and Astellas and received honoraria from Novartis, Celgene, and Incyte. J.A.P.-S. received honoraria and funding budget for research projects and is active on the advisory board and learning activities or conferences of Janssen, Takeda, Pfizer, Jazz, BMS, Amgen, and Gilead. J.F.S.-M. does consultancy and is a member of the board of directors advisory committees for AbbVie, Amgen, BMS, Celgene, GlaxoSmithKline (GSK), Janssen, Karyopharm, Merck Sharpe & Dohme, Novartis, Regeneron, Roche, Sanofi, SecuraBio, and Takeda. P.M. provides consultancy, is a member of the board of directors advisory committees and speaker’s bureau, and received research funding from Celgene, Sanofi, Incyte, Karyopharm, Novartis, Stemline/Menarini, Agios, Astellas Pharma, and Daiichi Sankyo; is also a member of the board of directors advisory committees for Pfizer, Teva, and AbbVie; received research funding from and is a member of speaker’s bureau for Janssen; and provides consultancy for Tolero Pharmaceutical, Forma Therapeutics, and Glycomimetics. B.P. served as a consultant and received honoraria from Adaptive, Amgen, BD Biosciences, BMS/Celgene, GSK, Janssen, Roche, Sanofi, and Takeda and received research support from BMS/Celgene, GSK, Roche, Sanofi, and Takeda. The remaining authors declare no competing financial interests.
Figures




References
-
- Weinberg OK, Pozdnyakova O, Campigotto F, et al. Reproducibility and prognostic significance of morphologic dysplasia in de novo acute myeloid leukemia. Mod Pathol. 2015;28(7):965–976. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

