GPC3-Unc5 receptor complex structure and role in cell migration
- PMID: 36240740
- PMCID: PMC9596381
- DOI: 10.1016/j.cell.2022.09.025
GPC3-Unc5 receptor complex structure and role in cell migration
Abstract
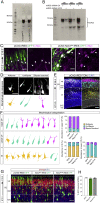
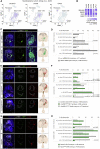
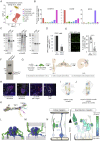
Neural migration is a critical step during brain development that requires the interactions of cell-surface guidance receptors. Cancer cells often hijack these mechanisms to disseminate. Here, we reveal crystal structures of Uncoordinated-5 receptor D (Unc5D) in complex with morphogen receptor glypican-3 (GPC3), forming an octameric glycoprotein complex. In the complex, four Unc5D molecules pack into an antiparallel bundle, flanked by four GPC3 molecules. Central glycan-glycan interactions are formed by N-linked glycans emanating from GPC3 (N241 in human) and C-mannosylated tryptophans of the Unc5D thrombospondin-like domains. MD simulations, mass spectrometry and structure-based mutants validate the crystallographic data. Anti-GPC3 nanobodies enhance or weaken Unc5-GPC3 binding and, together with mutant proteins, show that Unc5/GPC3 guide migrating pyramidal neurons in the mouse cortex, and cancer cells in an embryonic xenograft neuroblastoma model. The results demonstrate a conserved structural mechanism of cell guidance, where finely balanced Unc5-GPC3 interactions regulate cell migration.
Keywords: GPC3; UNC5A; UNC5B; UNC5C; UNC5D; Unc5; cell guidance; cell migration; cortex development; crystallography; glypican-3; nanobodies; neuroblastoma; stripe assay; structural biology; surface plasmon resonance; uncoordinated-5.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures












Comment in
-
Uncoordinated protein coordinates cell migration.Nature. 2022 Dec;612(7938):38-39. doi: 10.1038/d41586-022-03732-2. Nature. 2022. PMID: 36414730 No abstract available.
References
-
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/J.SOFTX.2015.06.001. - DOI
-
- ben Amar D., Thoinet K., Villalard B., Imbaud O., Costechareyre C., Jarrosson L., Reynaud F., Novion Ducassou J., Couté Y., Brunet J.-F., et al. Environmental cues from neural crest derivatives act as metastatic triggers in an embryonic neuroblastoma model. Nat. Commun. 2022;13:2549. doi: 10.1038/s41467-022-30237-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

