BefA, a microbiota-secreted membrane disrupter, disseminates to the pancreas and increases β cell mass
- PMID: 36240759
- PMCID: PMC9633563
- DOI: 10.1016/j.cmet.2022.09.001
BefA, a microbiota-secreted membrane disrupter, disseminates to the pancreas and increases β cell mass
Abstract
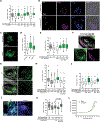
Microbiome dysbiosis is a feature of diabetes, but how microbial products influence insulin production is poorly understood. We report the mechanism of BefA, a microbiome-derived protein that increases proliferation of insulin-producing β cells during development in gnotobiotic zebrafish and mice. BefA disseminates systemically by multiple anatomic routes to act directly on pancreatic islets. We detail BefA's atomic structure, containing a lipid-binding SYLF domain, and demonstrate that it permeabilizes synthetic liposomes and bacterial membranes. A BefA mutant impaired in membrane disruption fails to expand β cells, whereas the pore-forming host defense protein, Reg3, stimulates β cell proliferation. Our work demonstrates that membrane permeabilization by microbiome-derived and host defense proteins is necessary and sufficient for β cell expansion during pancreas development, potentially connecting microbiome composition with diabetes risk.
Keywords: BefA; SYLF domain; diabetes; membrane permeabilization; microbiota; β cell proliferation.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.H.H. and K.G. are patent holders for the use of BefA, patent numbers 10563174, issued February 18, 2020, and 10968432, issued April 6, 2021.
Figures
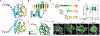


Comment in
-
Out of destruction comes new growth: Pore-forming antimicrobials make pancreas grow.Cell Metab. 2022 Nov 1;34(11):1611-1613. doi: 10.1016/j.cmet.2022.10.006. Cell Metab. 2022. PMID: 36323229
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography 66, 213–221. - PMC - PubMed
-
- Angelova MI, S.S., Méléard P, Faucon F, Bothorel P (1992). Preparation of giant vesicles by external AC electric fields. Kinetics and applications. In Trends in Colloid and Interface Science VI Progress in Colloid & Polymer Science, Helm C. LM, Möhwald H, ed. (Steinkopff; ).
-
- Astorri E, Guglielmi C, Bombardieri M, Alessandri C, Buzzetti R, Maggi D, Valesini G, Pitzalis C, and Pozzilli P (2010). Circulating Reg1alpha proteins and autoantibodies to Reg1alpha proteins as biomarkers of beta-cell regeneration and damage in type 1 diabetes. Horm Metab Res 42, 955–960. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

