Motor neurons use push-pull signals to direct vascular remodeling critical for their connectivity
- PMID: 36240771
- PMCID: PMC10316999
- DOI: 10.1016/j.neuron.2022.09.021
Motor neurons use push-pull signals to direct vascular remodeling critical for their connectivity
Abstract
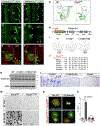
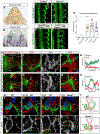
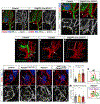
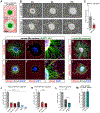
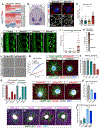
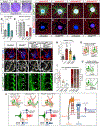
The nervous system requires metabolites and oxygen supplied by the neurovascular network, but this necessitates close apposition of neurons and endothelial cells. We find motor neurons attract vessels with long-range VEGF signaling, but endothelial cells in the axonal pathway are an obstacle for establishing connections with muscles. It is unclear how this paradoxical interference from heterotypic neurovascular contacts is averted. Through a mouse mutagenesis screen, we show that Plexin-D1 receptor is required in endothelial cells for development of neuromuscular connectivity. Motor neurons release Sema3C to elicit short-range repulsion via Plexin-D1, thus displacing endothelial cells that obstruct axon growth. When this signaling pathway is disrupted, epaxial motor neurons are blocked from reaching their muscle targets and concomitantly vascular patterning in the spinal cord is altered. Thus, an integrative system of opposing push-pull cues ensures detrimental axon-endothelial encounters are avoided while enabling vascularization within the nervous system and along peripheral nerves.
Keywords: VEGF; axon guidance; cell signaling; cell-cell interactions; forward genetics; motor neurons; neural circuits; neurovascular patterning; semaphorin/plexin; spinal cord.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Antipenko A, Himanen JP, van Leyen K, Nardi-Dei V, Lesniak J, Barton WA, Rajashankar KR, Lu M, Hoemme C, Püschel AW, et al. (2003). Structure of the semaphorin-3A receptor binding module. Neuron 39, 589–598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

