A-MYB/TCFL5 regulatory architecture ensures the production of pachytene piRNAs in placental mammals
- PMID: 36241367
- PMCID: PMC9808571
- DOI: 10.1261/rna.079472.122
A-MYB/TCFL5 regulatory architecture ensures the production of pachytene piRNAs in placental mammals
Abstract
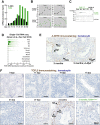
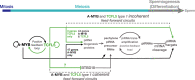
In male mice, the transcription factor A-MYB initiates the transcription of pachytene piRNA genes during meiosis. Here, we report that A-MYB activates the transcription factor Tcfl5 produced in pachytene spermatocytes. Subsequently, A-MYB and TCFL5 reciprocally reinforce their own transcription to establish a positive feedback circuit that triggers pachytene piRNA production. TCFL5 regulates the expression of genes required for piRNA maturation and promotes transcription of evolutionarily young pachytene piRNA genes, whereas A-MYB activates the transcription of older pachytene piRNA genes. Intriguingly, pachytene piRNAs from TCFL5-dependent young loci initiate the production of piRNAs from A-MYB-dependent older loci, ensuring the self-propagation of pachytene piRNAs. A-MYB and TCFL5 act via a set of incoherent feedforward loops that drive regulation of gene expression by pachytene piRNAs during spermatogenesis. This regulatory architecture is conserved in rhesus macaque, suggesting that it was present in the last common ancestor of placental mammals.
Keywords: A-MYB; TCFL5; pachytene piRNAs; spermatogenesis.
© 2023 Yu et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
