Low expression of EXOSC2 protects against clinical COVID-19 and impedes SARS-CoV-2 replication
- PMID: 36241425
- PMCID: PMC9585911
- DOI: 10.26508/lsa.202201449
Low expression of EXOSC2 protects against clinical COVID-19 and impedes SARS-CoV-2 replication
Abstract
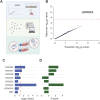
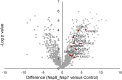
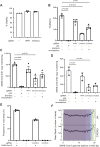
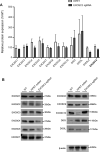
New therapeutic targets are a valuable resource for treatment of SARS-CoV-2 viral infection. Genome-wide association studies have identified risk loci associated with COVID-19, but many loci are associated with comorbidities and are not specific to host-virus interactions. Here, we identify and experimentally validate a link between reduced expression of EXOSC2 and reduced SARS-CoV-2 replication. EXOSC2 was one of the 332 host proteins examined, all of which interact directly with SARS-CoV-2 proteins. Aggregating COVID-19 genome-wide association studies statistics for gene-specific eQTLs revealed an association between increased expression of EXOSC2 and higher risk of clinical COVID-19. EXOSC2 interacts with Nsp8 which forms part of the viral RNA polymerase. EXOSC2 is a component of the RNA exosome, and here, LC-MS/MS analysis of protein pulldowns demonstrated interaction between the SARS-CoV-2 RNA polymerase and most of the human RNA exosome components. CRISPR/Cas9 introduction of nonsense mutations within EXOSC2 in Calu-3 cells reduced EXOSC2 protein expression and impeded SARS-CoV-2 replication without impacting cellular viability. Targeted depletion of EXOSC2 may be a safe and effective strategy to protect against clinical COVID-19.
© 2022 Moll et al.
Conflict of interest statement
MP Snyder is a co-founder and member of the scientific advisory board of Personalis, Qbio, January, SensOmics, Protos, Mirvie, NiMo, Onza and Oralome. He is also on the scientific advisory board of Danaher, Genapsys and Jupiter. The Krogan Laboratory has received research support from Vir Biotechnology, F Hoffmann-La Roche, and Rezo Therapeutics. Nevan Krogan has financially compensated consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics, Interline Therapeutics, Rezo Therapeutics, GEn1E Lifesciences, Inc. and Twist Bioscience Corp. He is on the Board of Directors of Rezo Therapeutics and is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, and Interline Therapeutics.
Figures


Update of
-
Low expression of EXOSC2 protects against clinical COVID-19 and impedes SARS-CoV-2 replication.bioRxiv [Preprint]. 2022 Mar 7:2022.03.06.483172. doi: 10.1101/2022.03.06.483172. bioRxiv. 2022. Update in: Life Sci Alliance. 2022 Oct 14;6(1):e202201449. doi: 10.26508/lsa.202201449. PMID: 35291294 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 216596/Z/19/Z/WT_/Wellcome Trust/United Kingdom
- S10 OD025212/OD/NIH HHS/United States
- MC_UU_00008/8/MRC_/Medical Research Council/United Kingdom
- IS-BRC-1215-20017/DH_/Department of Health/United Kingdom
- R01 HL101388/HL/NHLBI NIH HHS/United States
- P50 HL083800/HL/NHLBI NIH HHS/United States
- P30 DK116074/DK/NIDDK NIH HHS/United States
- R01 HL122939/HL/NHLBI NIH HHS/United States
- NF-SI-0617-10077/DH_/Department of Health/United Kingdom
- 203141/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- BB/S009566/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- UM1 HG009442/HG/NHGRI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
