Recent and future perspectives on engineering interferons and other cytokines as therapeutics
- PMID: 36241490
- PMCID: PMC9974544
- DOI: 10.1016/j.tibs.2022.09.005
Recent and future perspectives on engineering interferons and other cytokines as therapeutics
Abstract
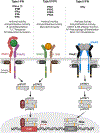
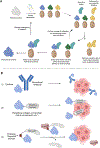
As crucial mediators and regulators of our immune system, cytokines are involved in a broad range of biological processes and are implicated in various disease pathologies. The field of cytokine therapeutics has gained much momentum from the maturation of conventional protein engineering methodologies such as structure-based designs and/or directed evolution, which is further aided by the advent of in silico protein designs and characterization. Just within the past 5 years, there has been an explosion of proof-of-concept, preclinical, and clinical studies that utilize an armory of protein engineering methods to develop cytokine-based drugs. Here, we highlight the key engineering strategies undertaken by recent studies that aim to improve the pharmacodynamic and pharmacokinetic profile of interferons and other cytokines as therapeutics.
Keywords: cytokines; interferons; interleukins; pleiotropy; protein engineering; therapeutics.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests J.L.M. is a co-inventor on patent applications US11198717B2, US20210309707A1, and US62/878,574, 2019, and has shares in Synthekine.
Figures
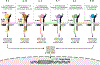


Similar articles
-
Engineering interferons and interleukins for cancer immunotherapy.Adv Drug Deliv Rev. 2022 Mar;182:114112. doi: 10.1016/j.addr.2022.114112. Epub 2022 Jan 24. Adv Drug Deliv Rev. 2022. PMID: 35085624 Review.
-
[Antitumoral action of interferons and interleukins in combination with radiotherapy. Part I: immunologic basis].Strahlenther Onkol. 2004 Apr;180(4):187-93. doi: 10.1007/s00066-004-9119-x. Strahlenther Onkol. 2004. PMID: 15057428 Review. German.
-
[Antitumoral action of interferons and interleukins in combination with radiotherapy. Part II: radiobiological and immunologic strategies].Strahlenther Onkol. 2004 Jun;180(6):331-9. doi: 10.1007/s00066-004-8119-1. Strahlenther Onkol. 2004. PMID: 15175867 Review. German.
-
Cytokines and their role as immunotherapeutics and vaccine Adjuvants: The emerging concepts.Cytokine. 2023 Sep;169:156268. doi: 10.1016/j.cyto.2023.156268. Epub 2023 Jun 13. Cytokine. 2023. PMID: 37320965 Review.
-
Cytokine-based therapy for metastatic renal cell cancer.Urol Clin North Am. 2003 Aug;30(3):589-600. doi: 10.1016/s0094-0143(03)00027-2. Urol Clin North Am. 2003. PMID: 12953757 Review.
Cited by
-
Interleukins in Platelet Biology: Unraveling the Complex Regulatory Network.Pharmaceuticals (Basel). 2024 Jan 13;17(1):109. doi: 10.3390/ph17010109. Pharmaceuticals (Basel). 2024. PMID: 38256942 Free PMC article. Review.
-
Advances and challenges in cancer immunotherapy: mechanisms, clinical applications, and future directions.Front Pharmacol. 2025 Jun 13;16:1602529. doi: 10.3389/fphar.2025.1602529. eCollection 2025. Front Pharmacol. 2025. PMID: 40584631 Free PMC article. Review.
-
Immunostimulatory CKb11 gene combined with immune checkpoint PD-1/PD-L1 blockade activates immune response and simultaneously overcomes the immunosuppression of cancer.Bioact Mater. 2024 May 23;39:239-254. doi: 10.1016/j.bioactmat.2024.05.014. eCollection 2024 Sep. Bioact Mater. 2024. PMID: 38832303 Free PMC article.
-
Liquid-phase separations coupled with ion mobility-mass spectrometry for next-generation biopharmaceutical analysis.Expert Rev Proteomics. 2024 May-Jun;21(5-6):259-270. doi: 10.1080/14789450.2024.2373707. Epub 2024 Jul 1. Expert Rev Proteomics. 2024. PMID: 38934922 Free PMC article. Review.
-
Structural studies of the IFNλ4 receptor complex using cryoEM enabled by protein engineering.Nat Commun. 2025 Jan 18;16(1):818. doi: 10.1038/s41467-025-56119-y. Nat Commun. 2025. PMID: 39827213 Free PMC article.
References
-
- Oppenheim JJ, Cytokines: past, present, and future. Int J Hematol, 2001. 74(1): p. 3–8. - PubMed
-
- Turner MD, et al., Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2014. 1843(11): p. 2563–2582. - PubMed
-
- Ait-Oufella H, et al., Recent Advances on the Role of Cytokines in Atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology, 2011. 31(5): p. 969–979. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

