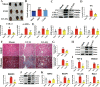
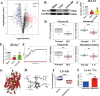
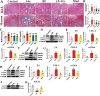
Fig. 6. The anti-fibrotic effects of Mad in UUO model.
A Picture of left kidneys of mice with different treatments (n = 3). B The renal index (mg/g) (n = 3). *p = 0.0140 for B: Sham vs. UUO, p > 0.9999 for B: Sham vs. Mad (p.o), p = 0.7027 for B: Sham vs. Mad (i.p), #p = 0.0175 for B: UUO vs. Mad (p.o), p = 0.1476 for B: UUO vs. Mad (i.p). C Representative photomicrographs of the H&E staining from left kidneys of Sham, UUO, Mad (p.o.) or Mad (i.p.)-treated UUO mice. D Bar graphs depict renal injury scores based on H&E staining (n = 3). ***p = 0.0003 for injury score: Sham vs. UUO, p = 0.1712 for injury score: Sham vs. Mad (p.o), ***p = 0.0010 for injury score: Sham vs. Mad (i.p), ##p = 0.0047 for injury score: UUO vs. Mad (p.o), p = 0.8302 for injury score: UUO vs. Mad (i.p). E Representative photomicrographs of the H&E staining and Masson’s trichrome staining from left kidneys of Sham, UUO, UUO + MA, UUO + Mad (p.o.), UUO + Mad + Antibiotic mice (H&E and Masson’s staining; scale bar, 100 μm; magnification, ×200). F Bar graphs depict renal injury scores and renal interstitial fibrosis scores based on H&E staining or Masson’s trichrome staining (n = 3). ***p = 0.0002 for injury score: Sham vs. UUO, ***p = 0.0002 for injury score: Sham vs. UUO + MA, p = 0.0692 for injury score: Sham vs. UUO + Mad, ***p = 0.0003 for injury score: Sham vs. Mad+Antibiotic, p > 0.9999 for injury score: UUO vs. UUO + MA, #p = 0.0119 for injury score: UUO vs. UUO + Mad, p = 0.9984 for injury score: UUO vs. Mad+Antibiotic; ***p = 0.0006 for fibrosis score: Sham vs. UUO, ***p = 0.0006 for fibrosis score: Sham vs. UUO + MA, p = 0.0687 for fibrosis score: Sham vs. UUO + Mad, ***p = 0.0006 for fibrosis score: Sham vs. Mad+Antibiotic, p > 0.9999 for fibrosis score: UUO vs. UUO + MA, #p = 0.0101 for fibrosis score: UUO vs. UUO + Mad, p > 0.9999 for fibrosis score: UUO vs. Mad+Antibiotic. G Biochemical parameters including BUN, Scr, TC, TG in each of mice (n = 6). ***p < 0.0001 for BUN: Sham vs. UUO, ***p < 0.0001 for BUN: Sham vs. UUO + MA, p = 0.7803 for BUN: Sham vs. UUO + Mad, ***p < 0.0001 for BUN: Sham vs. Mad+Antibiotic, p = 0.9949 for BUN: UUO vs. UUO + MA, ###p < 0.0001 for BUN: UUO vs. UUO + Mad, p = 0.9366 for BUN: UUO vs. Mad+Antibiotic; ***p < 0.0001 for SCR: Sham vs. UUO, ***p < 0.0001 for SCR: Sham vs. UUO + MA, **p = 0.0074 for SCR: Sham vs. UUO + Mad, ***p < 0.0001 for SCR: Sham vs. Mad+Antibiotic, p > 0.9999 for SCR: UUO vs. UUO + MA, ###p = 0.0005 for SCR: UUO vs. UUO + Mad, p = 0.9982 for SCR: UUO vs. Mad+Antibiotic; ***p < 0.0001 for TC: Sham vs. UUO, ***p < 0.0001 for TC: Sham vs. UUO + MA, p = 0.9957 for TC: Sham vs. UUO + Mad, ***p = 0.0001 for TC: Sham vs. Mad+Antibiotic, p > 0.9999 for TC: UUO vs. UUO + MA, ###p = 0.0003 for TC: UUO vs. UUO + Mad, p > 0.9999 for TC: UUO vs. Mad+Antibiotic; ***p < 0.0001 for TG: Sham vs. UUO, ***p < 0.0001 for TG: Sham vs. UUO + MA, p = 0.1726 for TG: Sham vs. UUO + Mad, ***p < 0.0001 for TG: Sham vs. Mad+Antibiotic, p = 0.9997 for TG: UUO vs. UUO + MA, ###p < 0.0001 for TG: UUO vs. UUO + Mad, p = 0.9810 for TG: UUO vs. Mad+Antibiotic. H Kidney expression of FN, Col I and α-SMA from all groups, assayed by Western blot. I Quantitative analysis of Fig. 6H (n = 3). ***p = 0.0001 for FN: Sham vs. UUO, ***p = 0.0002 for FN: Sham vs. UUO + MA, p = 0.8340 for FN: Sham vs. UUO + Mad, ***p < 0.0001 for FN: Sham vs. Mad+Antibiotic, p > 0.9999 for FN: UUO vs. UUO + MA, ###p = 0.0006 for FN: UUO vs. UUO + Mad, p = 0.9149 for FN: UUO vs. Mad+Antibiotic; *p = 0.0133 for COL-1: Sham vs. UUO, *p = 0.0240 for COL-1: Sham vs. UUO + MA, p > 0.9999 for COL-1: Sham vs. UUO + Mad, **p = 0.0086 for COL-1: Sham vs. Mad+Antibiotic, p > 0.9999 for COL-1: UUO vs. UUO + MA, #p = 0.0139 for COL-1: UUO vs. UUO + Mad, p > 0.9999 for COL-1: UUO vs. Mad+Antibiotic; **p = 0.0079 for α-SMA: Sham vs. UUO, *p = 0.0480 for α-SMA: Sham vs. UUO + MA, p = 0.9946 for α-SMA: Sham vs. UUO + Mad, p = 0.0607 for α-SMA: Sham vs. Mad+Antibiotic, p = 0.9054 for α-SMA: UUO vs. UUO + MA, #p = 0.0223 for α-SMA: UUO vs. UUO + Mad, p = 0.8440 for α-SMA: UUO vs. Mad+Antibiotic. J Kidney expression of TGF-β/Smad signaling pathway from Sham, UUO, Mad-treated UUO mice, assayed by Western blot. K Quantitative analysis of Fig. 6J (n = 3). **p = 0.0050 for TGF-β: Sham vs. UUO, #p = 0.0428 for TGF-β: UUO vs. UUO + Mad; ***p < 0.0001 for SMAD2: Sham vs. UUO, *p = 0.0173 for SMAD2: Sham vs. UUO + Mad, ###p < 0.0001 for SMAD2: UUO vs. UUO + Mad; **p = 0.0018 for SMAD3: Sham vs. UUO, p = 0.2011 for SMAD3: Sham vs. UUO + Mad, #p = 0.0135 for SMAD3: UUO vs. UUO+Mad. L Representative Western blot of Nrf2, keap1, 12-LOX, Rac-1. M Quantitative analysis of Fig. 6L (n = 3). *p = 0.0271 for NRF2: Sham vs. UUO, p = 0.7328 for NRF2: Sham vs. UUO + Mad, ##p = 0.0091 for NRF2: UUO vs. UUO + Mad; **p = 0.0083 for KEAP1: Sham vs. UUO, #p = 0.0351 for KEAP1: UUO vs. UUO + Mad; ***p = 0.0001 for 12-LOX: Sham vs. UUO, p = 0.3690 for 12-LOX: Sham vs. UUO + Mad, ###p = 0.0003 for 12-LOX: UUO vs. UUO + Mad; **p = 0.0065 for RAC1: Sham vs. UUO, p = 0.4954 for RAC1: Sham vs. UUO + Mad, #p = 0.0300 for RAC1: UUO vs. UUO + Mad. N The relative abundance of B. fragilis in the sham, UUO and UUO + Mad groups measured by qPCR (n = 3). *p = 0.0309 for N: Sham vs. UUO, #p = 0.0122 for N: UUO vs. UUO + Mad. Data are presented as mean ± SD. Comparison in N were performed with a two-tailed Student’s t test. Comparisons in B, D, F, G, I, K, M and N were compared using One-Way ANOVA followed by Sidak’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001 (compared with sham group), #P < 0.05, ##P < 0.01, ###P < 0.001 (compared with UUO group). Individual data points are independent biological replicates unless otherwise stated.