Phosphatase protector alpha4 (α4) is involved in adipocyte maintenance and mitochondrial homeostasis through regulation of insulin signaling
- PMID: 36241662
- PMCID: PMC9568526
- DOI: 10.1038/s41467-022-33842-4
Phosphatase protector alpha4 (α4) is involved in adipocyte maintenance and mitochondrial homeostasis through regulation of insulin signaling
Abstract
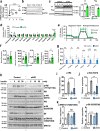
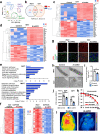
Insulin signaling is mediated via a network of protein phosphorylation. Dysregulation of this network is central to obesity, type 2 diabetes and metabolic syndrome. Here we investigate the role of phosphatase binding protein Alpha4 (α4) that is essential for the serine/threonine protein phosphatase 2A (PP2A) in insulin action/resistance in adipocytes. Unexpectedly, adipocyte-specific inactivation of α4 impairs insulin-induced Akt-mediated serine/threonine phosphorylation despite a decrease in the protein phosphatase 2A (PP2A) levels. Interestingly, loss of α4 also reduces insulin-induced insulin receptor tyrosine phosphorylation. This occurs through decreased association of α4 with Y-box protein 1, resulting in the enhancement of the tyrosine phosphatase protein tyrosine phosphatase 1B (PTP1B) expression. Moreover, adipocyte-specific knockout of α4 in male mice results in impaired adipogenesis and altered mitochondrial oxidation leading to increased inflammation, systemic insulin resistance, hepatosteatosis, islet hyperplasia, and impaired thermogenesis. Thus, the α4 /Y-box protein 1(YBX1)-mediated pathway of insulin receptor signaling is involved in maintaining insulin sensitivity, normal adipose tissue homeostasis and systemic metabolism.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

