Investigation of radiomics based intra-patient inter-tumor heterogeneity and the impact of tumor subsampling strategies
- PMID: 36241749
- PMCID: PMC9568579
- DOI: 10.1038/s41598-022-20931-z
Investigation of radiomics based intra-patient inter-tumor heterogeneity and the impact of tumor subsampling strategies
Abstract
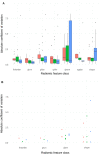
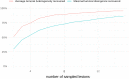
While radiomics analysis has been applied for localized cancer disease, its application to the metastatic setting involves a non-exhaustive lesion subsampling strategy which may sidestep the intrapatient tumoral heterogeneity, hindering the reproducibility and the therapeutic response performance. Our aim was to evaluate if radiomics features can capture intertumoral intrapatient heterogeneity, and the impact of tumor subsampling on the computed heterogeneity. To this end, We delineated and extracted radiomics features of all visible tumors from single acquisition pre-treatment computed tomography of patients with metastatic lung cancer (cohort L) and confirmed our results on a larger cohort of patients with metastatic melanoma (cohort M). To quantify the captured heterogeneity, the absolute coefficient of variation (CV) of each radiomics index was calculated at the patient-level and a sensitivity analysis was performed using only a subset of all extracted features robust to the segmentation step. The extent of information loss by six commonly used tumor sampling strategies was then assessed. A total of 602 lesions were segmented from 43 patients (median age 57, 4.9% female). All robust radiomics indexes exhibited at least 20% of variation with significant heterogeneity both in heavily and oligo metastasized patients, and also at the organ level. None of the segmentation subsampling strategies were able to recover the true tumoral heterogeneity obtained by exhaustive tumor sampling. Image-based inter-tumor intra-patient heterogeneity can be successfully grasped by radiomics analyses. Failing to take into account this kind of heterogeneity will lead to inconsistent predictive algorithms. Guidelines to standardize the tumor sampling step and/or AI-driven tools to alleviate the segmentation effort are required.
© 2022. The Author(s).
Conflict of interest statement
R.S. reports research and travel grants from Fondation ARC and Université Paris-Saclay and support from Inserm and Fondation Bettencourt Schueller outside of the submitted work. Ca.R. is a consultant advisor for Amgen, AstraZeneca, BMS, Merck, MSD, Novartis, Pierre Fabre, Roche and Sanofi. E.D. reports grants and personal fees from Roche Genentech, grants from Servier, grants from Astrazeneca, grants and personal fees from Merck Serono, grants from BMS, and grants from MSD, outside the submitted work. All remaining authors have declared no conflicts of interest.
Figures




Similar articles
-
Metastatic Lung Adenocarcinomas: Development and Evaluation of Radiomic-Based Methods to Measure Baseline Intra-Patient Inter-Tumor Lesion Heterogeneity.J Imaging Inform Med. 2025 Feb;38(1):148-164. doi: 10.1007/s10278-024-01163-1. Epub 2024 Jul 17. J Imaging Inform Med. 2025. PMID: 39020153 Free PMC article.
-
Lung tumor segmentation methods: Impact on the uncertainty of radiomics features for non-small cell lung cancer.PLoS One. 2018 Oct 4;13(10):e0205003. doi: 10.1371/journal.pone.0205003. eCollection 2018. PLoS One. 2018. PMID: 30286184 Free PMC article.
-
Radiomic signature based on CT imaging to distinguish invasive adenocarcinoma from minimally invasive adenocarcinoma in pure ground-glass nodules with pleural contact.Cancer Imaging. 2021 Jan 6;21(1):1. doi: 10.1186/s40644-020-00376-1. Cancer Imaging. 2021. PMID: 33407884 Free PMC article.
-
Robust imaging habitat computation using voxel-wise radiomics features.Sci Rep. 2021 Oct 11;11(1):20133. doi: 10.1038/s41598-021-99701-2. Sci Rep. 2021. PMID: 34635786 Free PMC article.
-
Radiomics and Radiogenomics of Ovarian Cancer: Implications for Treatment Monitoring and Clinical Management.Radiol Clin North Am. 2023 Jul;61(4):749-760. doi: 10.1016/j.rcl.2023.02.006. Epub 2023 Mar 31. Radiol Clin North Am. 2023. PMID: 37169435 Review.
Cited by
-
Textural heterogeneity of liver lesions in CT imaging - comparison of colorectal and pancreatic metastases.Abdom Radiol (NY). 2024 Dec;49(12):4295-4306. doi: 10.1007/s00261-024-04511-5. Epub 2024 Aug 8. Abdom Radiol (NY). 2024. PMID: 39115682 Free PMC article.
-
Measured intrapatient radiomic variability as a predictor of treatment response in multi-metastatic soft tissue sarcoma patients.medRxiv [Preprint]. 2025 May 14:2025.04.11.25325700. doi: 10.1101/2025.04.11.25325700. medRxiv. 2025. Update in: Sci Rep. 2025 Jul 30;15(1):27838. doi: 10.1038/s41598-025-12451-3. PMID: 40297462 Free PMC article. Updated. Preprint.
-
18F-FDG PET/CT Radiomics for Predicting Therapy Response in Primary Mediastinal B-Cell Lymphoma: A Bi-Centric Pilot Study.Cancers (Basel). 2025 May 30;17(11):1827. doi: 10.3390/cancers17111827. Cancers (Basel). 2025. PMID: 40507310 Free PMC article.
-
Radiomic analysis of patient and interorgan heterogeneity in response to immunotherapies and BRAF-targeted therapy in metastatic melanoma.J Immunother Cancer. 2025 Feb 12;13(2):e009568. doi: 10.1136/jitc-2024-009568. J Immunother Cancer. 2025. PMID: 39939139 Free PMC article.
-
Radiomic-Based Prediction of Lesion-Specific Systemic Treatment Response in Metastatic Disease.medRxiv [Preprint]. 2024 Aug 13:2023.09.22.23294942. doi: 10.1101/2023.09.22.23294942. medRxiv. 2024. Update in: Comput Med Imaging Graph. 2024 Sep;116:102413. doi: 10.1016/j.compmedimag.2024.102413. PMID: 37873411 Free PMC article. Updated. Preprint.

