Metagenomic analysis of viromes in tissues of wild Qinghai vole from the eastern Tibetan Plateau
- PMID: 36241909
- PMCID: PMC9562062
- DOI: 10.1038/s41598-022-22134-y
Metagenomic analysis of viromes in tissues of wild Qinghai vole from the eastern Tibetan Plateau
Abstract
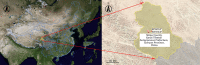
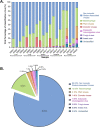
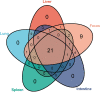
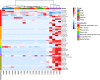
Rodents are natural reservoirs of diverse zoonotic viruses and widely distributed on the Tibetan Plateau. A comprehensive understanding of the virome in local rodent species could provide baseline of viral content and assist in efforts to reduce the risk for future emergence of rodent related zoonotic diseases. A total of 205 tissue and fecal samples from 41 wild Qinghai voles were collected. Metagenomic analyses were performed to outline the characteristics of the viromes, and phylogenetic analyses were used to identify the novel viral genomes. The virome distribution among five tissues (liver, lung, spleen, small intestine with content and feces) was also compared. We identified sequences related to 46 viral families. Novel viral genomes from distinct evolutionary lineages with known viruses were characterized for their genomic and evolutionary characteristics, including Hepatovirus, Hepacivirus, Rotavirus, and Picobirnavirus. Further analyses revealed that the core virome harbored by rodent internal tissues were quite different from the virome found in intestine and fecal samples. These findings provide an overview of the viromes in wild Qinghai voles, which are unique and the most common rodent species in the eastern Tibetan Plateau. A high diversity of viruses is likely present in rodent species in this area.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Decoding the RNA viromes in rodent lungs provides new insight into the origin and evolutionary patterns of rodent-borne pathogens in Mainland Southeast Asia.Microbiome. 2021 Jan 21;9(1):18. doi: 10.1186/s40168-020-00965-z. Microbiome. 2021. PMID: 33478588 Free PMC article.
-
Gut Virome of the World's Highest-Elevation Lizard Species (Phrynocephalus erythrurus and Phrynocephalus theobaldi) Reveals Versatile Commensal Viruses.Microbiol Spectr. 2022 Feb 23;10(1):e0187221. doi: 10.1128/spectrum.01872-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196818 Free PMC article.
-
Metagenomic analysis of viral diversity and a novel astroviruse of forest rodent.Virol J. 2022 Aug 31;19(1):138. doi: 10.1186/s12985-022-01847-6. Virol J. 2022. PMID: 36045380 Free PMC article.
-
Diversity, evolution, and emergence of fish viruses.J Virol. 2024 Jun 13;98(6):e0011824. doi: 10.1128/jvi.00118-24. Epub 2024 May 24. J Virol. 2024. PMID: 38785422 Free PMC article. Review.
-
Zoonotic disease and virome diversity in bats.Curr Opin Virol. 2022 Feb;52:192-202. doi: 10.1016/j.coviro.2021.12.008. Epub 2021 Dec 23. Curr Opin Virol. 2022. PMID: 34954661 Free PMC article. Review.
Cited by
-
Evaluating the zoonotic potential of RNA viromes of rodents provides new insight into rodent-borne zoonotic pathogens in Guangdong, China.One Health. 2023 Sep 20;17:100631. doi: 10.1016/j.onehlt.2023.100631. eCollection 2023 Dec. One Health. 2023. PMID: 38024253 Free PMC article.
-
Virome characterization of field-collected rodents in suburban city.NPJ Biofilms Microbiomes. 2025 Jun 11;11(1):103. doi: 10.1038/s41522-025-00740-8. NPJ Biofilms Microbiomes. 2025. PMID: 40500267 Free PMC article.
-
The Genetic Diversity and Interspecific Transmission of Circovirus in Rhizomys sinensis in Guangdong, Southern China.Transbound Emerg Dis. 2023 Nov 9;2023:6668569. doi: 10.1155/2023/6668569. eCollection 2023. Transbound Emerg Dis. 2023. PMID: 40303771 Free PMC article.
-
Multiple infections of zoonotic pathogens in wild Brandt's voles (Lasiopodomys brandtii).Vet Med Sci. 2023 Sep;9(5):2201-2211. doi: 10.1002/vms3.1214. Epub 2023 Jul 25. Vet Med Sci. 2023. PMID: 37491010 Free PMC article.
-
The silent spread: uncovering the diversity and evolution of poxviruses in ticks across Western China's host landscapes.Virol J. 2025 Jun 30;22(1):214. doi: 10.1186/s12985-025-02844-1. Virol J. 2025. PMID: 40588758 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

