DAZAP1 facilitates the alternative splicing of KITLG to promote multiple myeloma cell proliferation via ERK signaling pathway
- PMID: 36242590
- PMCID: PMC9596219
- DOI: 10.18632/aging.204326
DAZAP1 facilitates the alternative splicing of KITLG to promote multiple myeloma cell proliferation via ERK signaling pathway
Abstract
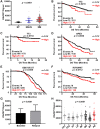
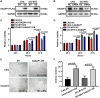
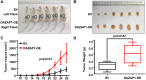
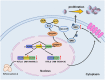
Multiple myeloma (MM) is an incurable plasma cell malignancy, in which alternative pre-mRNA splicing (AS) acts as one of the key transcriptome modifier. The Deleted in Azoospermia-Associated Protein 1 (DAZAP1) is a splicing factor that has been identified as an oncogene in multiple cancers, yet its role in MM proliferation remains unclear. We first analyzed MM clinical databases and found that MM patients with elevated DAZAP1 had a poor survival. Furthermore, we overexpressed DAZAP1 by lentiviral transfection and utilized siRNA silencing the expression of DAZAP1 in MM cells. DAZAP1 promoted MM cell proliferation in vitro and accelerated MM xenograft tumor growth in vivo. KEGG pathway enrichment analysis showed that ERK signaling pathway was activated in DAZAP1-OE MM cells. The analyses of RIP-seq and RIP-qPCR revealed that DAZAP1 activated alternative splicing of KIT proto-oncogene ligand (KITLG) mRNA. Further study validated that DAZAP1 increased ERK phosphorylation via modulating alternative splicing of KITLG mRNA to promote MM cell proliferation. In conclusion, we establish DAZAP1 as a tumor-promoting gene with therapeutic potential and provide mechanistic insights into targeting DAZAP1 as a new strategy for the diagnosis and treatment of MM.
Keywords: DAZAP1; ERK; KITLG; alternative splicing; multiple myeloma.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

