Nociceptor neurons direct goblet cells via a CGRP-RAMP1 axis to drive mucus production and gut barrier protection
- PMID: 36243004
- PMCID: PMC9617795
- DOI: 10.1016/j.cell.2022.09.024
Nociceptor neurons direct goblet cells via a CGRP-RAMP1 axis to drive mucus production and gut barrier protection
Abstract
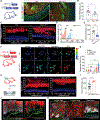
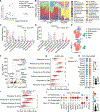
Neuroepithelial crosstalk is critical for gut physiology. However, the mechanisms by which sensory neurons communicate with epithelial cells to mediate gut barrier protection at homeostasis and during inflammation are not well understood. Here, we find that Nav1.8+CGRP+ nociceptor neurons are juxtaposed with and signal to intestinal goblet cells to drive mucus secretion and gut protection. Nociceptor ablation led to decreased mucus thickness and dysbiosis, while chemogenetic nociceptor activation or capsaicin treatment induced mucus growth. Mouse and human goblet cells expressed Ramp1, receptor for the neuropeptide CGRP. Nociceptors signal via the CGRP-Ramp1 pathway to induce rapid goblet cell emptying and mucus secretion. Notably, commensal microbes activated nociceptors to control homeostatic CGRP release. In the absence of nociceptors or epithelial Ramp1, mice showed increased epithelial stress and susceptibility to colitis. Conversely, CGRP administration protected nociceptor-ablated mice against colitis. Our findings demonstrate a neuron-goblet cell axis that orchestrates gut mucosal barrier protection.
Keywords: CGRP; Ramp1; colitis; goblet cell; gut barrier; gut-brain axis; mucus; neuroepithelial crosstalk; nociceptor; sensory neuron.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.J. is an employee of Genentech, Inc. I.M.C. serves on scientific advisory boards of GSK Pharmaceuticals and Limm Therapeutics. His lab receives research support from Moderna Inc. and AbbVie/Allergan Pharmaceuticals.
Figures





Comment in
-
Adding data to gut feelings.Cell. 2022 Oct 27;185(22):4046-4048. doi: 10.1016/j.cell.2022.10.002. Cell. 2022. PMID: 36306732
-
Pain-sensing neurons calm the gut.Nat Rev Immunol. 2022 Dec;22(12):716. doi: 10.1038/s41577-022-00808-y. Nat Rev Immunol. 2022. PMID: 36357704 No abstract available.
-
Gutsy sensations modulate intestinal disease.Trends Immunol. 2023 Jan;44(1):1-3. doi: 10.1016/j.it.2022.11.007. Epub 2022 Dec 8. Trends Immunol. 2023. PMID: 36503685
-
A triplex for intestinal protection: Neurons, microbes, and goblet cells.Immunity. 2022 Dec 13;55(12):2228-2230. doi: 10.1016/j.immuni.2022.11.006. Immunity. 2022. PMID: 36516817
-
How pain sensors make the gut weep.Cell Res. 2023 May;33(5):339-340. doi: 10.1038/s41422-022-00768-x. Cell Res. 2023. PMID: 36627347 Free PMC article. No abstract available.
References
-
- Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP, Nassar MA, Dickenson AH, and Wood JN (2008). The cell and molecular basis of mechanical, cold, and inflammatory pain. Science 321, 702–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

