Comparative genome analysis of mycobacteria focusing on tRNA and non-coding RNA
- PMID: 36243697
- PMCID: PMC9569102
- DOI: 10.1186/s12864-022-08927-5
Comparative genome analysis of mycobacteria focusing on tRNA and non-coding RNA
Abstract
Background: The Mycobacterium genus encompasses at least 192 named species, many of which cause severe diseases such as tuberculosis. Non-tuberculosis mycobacteria (NTM) can also infect humans and animals. Some are of emerging concern because they show high resistance to commonly used antibiotics while others are used and evaluated in bioremediation or included in anticancer vaccines.
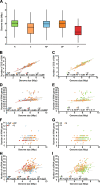
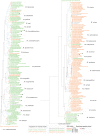
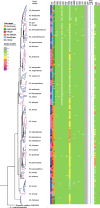
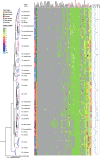
Results: We provide the genome sequences for 114 mycobacterial type strains and together with 130 available mycobacterial genomes we generated a phylogenetic tree based on 387 core genes and supported by average nucleotide identity (ANI) data. The 244 genome sequences cover most of the species constituting the Mycobacterium genus. The genome sizes ranged from 3.2 to 8.1 Mb with an average of 5.7 Mb, and we identified 14 new plasmids. Moreover, mycobacterial genomes consisted of phage-like sequences ranging between 0 and 4.64% dependent on mycobacteria while the number of IS elements varied between 1 and 290. Our data also revealed that, depending on the mycobacteria, the number of tRNA and non-coding (nc) RNA genes differ and that their positions on the chromosome varied. We identified a conserved core set of 12 ncRNAs, 43 tRNAs and 18 aminoacyl-tRNA synthetases among mycobacteria.
Conclusions: Phages, IS elements, tRNA and ncRNAs appear to have contributed to the evolution of the Mycobacterium genus where several tRNA and ncRNA genes have been horizontally transferred. On the basis of our phylogenetic analysis, we identified several isolates of unnamed species as new mycobacterial species or strains of known mycobacteria. The predicted number of coding sequences correlates with genome size while the number of tRNA, rRNA and ncRNA genes does not. Together these findings expand our insight into the evolution of the Mycobacterium genus and as such they establish a platform to understand mycobacterial pathogenicity, their evolution, antibiotic resistance/tolerance as well as the function and evolution of ncRNA among mycobacteria.
Keywords: Core gene phylogeny; Mycobacterial genomes; tRNA and non-coding RNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K, Ludwig W, Whitman WB. Bergey's manual of systematic bacteriology. 2. New York: Springer, New York; 2012.
-
- Hatfull GF, Jacobs WR. Molecular genetics of mycobacteria, second edition. Washington, DC: ASM press; 2014.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

