Co-expression network analysis identifies potential candidate hub genes in severe influenza patients needing invasive mechanical ventilation
- PMID: 36243706
- PMCID: PMC9569050
- DOI: 10.1186/s12864-022-08915-9
Co-expression network analysis identifies potential candidate hub genes in severe influenza patients needing invasive mechanical ventilation
Abstract
Background: Influenza is a contagious disease that affects people of all ages and is linked to considerable mortality during epidemics and occasional outbreaks. Moreover, effective immunological biomarkers are needed for elucidating aetiology and preventing and treating severe influenza. Herein, we aimed to evaluate the key genes linked with the disease severity in influenza patients needing invasive mechanical ventilation (IMV). Three gene microarray data sets (GSE101702, GSE21802, and GSE111368) from blood samples of influenza patients were made available by the Gene Expression Omnibus (GEO) database. The GSE101702 and GSE21802 data sets were combined to create the training set. Hub indicators for IMV patients with severe influenza were determined using differential expression analysis and Weighted correlation network analysis (WGCNA) from the training set. The receiver operating characteristic curve (ROC) was also used to evaluate the hub genes from the test set's diagnostic accuracy. Different immune cells' infiltration levels in the expression profile and their correlation with hub gene markers were examined using single-sample gene set enrichment analysis (ssGSEA).
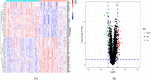
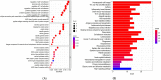
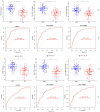
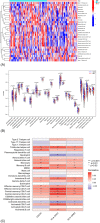
Results: In the present study, we evaluated a total of 447 differential genes. WGCNA identified eight co-expression modules, with the red module having the strongest correlation with IMV patients. Differential genes were combined to obtain 3 hub genes (HLA-DPA1, HLA-DRB3, and CECR1). The identified genes were investigated as potential indicators for patients with severe influenza who required IMV using the least absolute shrinkage and selection operator (LASSO) approach. The ROC showed the diagnostic value of the three hub genes in determining the severity of influenza. Using ssGSEA, it has been revealed that the expression of key genes was negatively correlated with neutrophil activation and positively associated with adaptive cellular immune response.
Conclusion: We evaluated three novel hub genes that could be linked to the immunopathological mechanism of severe influenza patients who require IMV treatment and could be used as potential biomarkers for severe influenza prevention and treatment.
Keywords: Co-expression network analysis; Hub gene; Invasive mechanical ventilation; Severe influenza.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Bioinformatics analysis identifies a key gene HLA_DPA1 in severe influenza-associated immune infiltration.BMC Genomics. 2024 Mar 7;25(1):257. doi: 10.1186/s12864-024-10184-7. BMC Genomics. 2024. PMID: 38454348 Free PMC article.
-
Dysregulation and imbalance of innate and adaptive immunity are involved in the cardiomyopathy progression.Front Cardiovasc Med. 2022 Sep 6;9:973279. doi: 10.3389/fcvm.2022.973279. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36148059 Free PMC article.
-
Genetic analysis of cuproptosis subtypes and immunological features in severe influenza.Microb Pathog. 2023 Jul;180:106162. doi: 10.1016/j.micpath.2023.106162. Epub 2023 May 18. Microb Pathog. 2023. PMID: 37207785
-
Identification of Hub Biomarkers and Immune-Related Pathways Participating in the Progression of Antineutrophil Cytoplasmic Antibody-Associated Glomerulonephritis.Front Immunol. 2022 Jan 5;12:809325. doi: 10.3389/fimmu.2021.809325. eCollection 2021. Front Immunol. 2022. PMID: 35069594 Free PMC article.
-
Identification of key candidate biomarkers for severe influenza infection by integrated bioinformatical analysis and initial clinical validation.J Cell Mol Med. 2021 Feb;25(3):1725-1738. doi: 10.1111/jcmm.16275. Epub 2021 Jan 14. J Cell Mol Med. 2021. PMID: 33448094 Free PMC article.
Cited by
-
Linked Mutations in the Ebola Virus Polymerase Are Associated with Organ Specific Phenotypes.Microbiol Spectr. 2023 Mar 22;11(2):e0415422. doi: 10.1128/spectrum.04154-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36946725 Free PMC article.
-
Development and validation of a nomogram to predict severe influenza.Immun Inflamm Dis. 2024 Sep;12(9):e70026. doi: 10.1002/iid3.70026. Immun Inflamm Dis. 2024. PMID: 39340342 Free PMC article.
-
Prevalence of neutropenia in US residents: a population based analysis of NHANES 2011-2018.BMC Public Health. 2023 Jun 28;23(1):1254. doi: 10.1186/s12889-023-16141-5. BMC Public Health. 2023. PMID: 37380948 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

