Discriminating cross-reactivity in polyclonal IgG1 responses against SARS-CoV-2 variants of concern
- PMID: 36243713
- PMCID: PMC9568977
- DOI: 10.1038/s41467-022-33899-1
Discriminating cross-reactivity in polyclonal IgG1 responses against SARS-CoV-2 variants of concern
Abstract
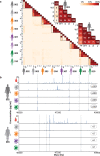
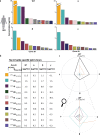
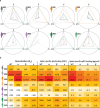
Existing assays to measure antibody cross-reactivity against different SARS-CoV-2 spike (S) protein variants lack the discriminatory power to provide insights at the level of individual clones. Using a mass spectrometry-based approach we are able to monitor individual donors' IgG1 clonal responses following a SARS-CoV-2 infection. We monitor the plasma clonal IgG1 profiles of 8 donors who had experienced an infection by either the wild type Wuhan Hu-1 virus or one of 3 VOCs (Alpha, Beta and Gamma). In these donors we chart the full plasma IgG1 repertoires as well as the IgG1 repertoires targeting the SARS-CoV-2 spike protein trimer VOC antigens. The plasma of each donor contains numerous anti-spike IgG1 antibodies, accounting for <0.1% up to almost 10% of all IgG1s. Some of these antibodies are VOC-specific whereas others do recognize multiple or even all VOCs. We show that in these polyclonal responses, each clone exhibits a distinct cross-reactivity and also distinct virus neutralization capacity. These observations support the need for a more personalized look at the antibody clonal responses to infectious diseases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- WHO. Tracking SARS-CoV-2 variants, https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (2022).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

