Features and factors that dictate if terminating ribosomes cause or counteract nonsense-mediated mRNA decay
- PMID: 36244451
- PMCID: PMC9661723
- DOI: 10.1016/j.jbc.2022.102592
Features and factors that dictate if terminating ribosomes cause or counteract nonsense-mediated mRNA decay
Abstract
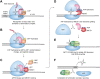
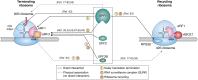
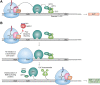
Nonsense-mediated mRNA decay (NMD) is a quality control pathway in eukaryotes that continuously monitors mRNA transcripts to ensure truncated polypeptides are not produced. The expression of many normal mRNAs that encode full-length polypeptides is also regulated by this pathway. Such transcript surveillance by NMD is intimately linked to translation termination. When a ribosome terminates translation at a normal termination codon, NMD is not activated, and mRNA can undergo repeated rounds of translation. On the other hand, when translation termination is deemed abnormal, such as that on a premature termination codon, it leads to a series of poorly understood events involving the NMD pathway, which destabilizes the transcript. In this review, we summarize our current understanding of how the NMD machinery interfaces with the translation termination factors to initiate NMD. We also discuss a variety of cis-acting sequence contexts and trans-acting factors that can cause readthrough, ribosome reinitiation, or ribosome frameshifting at stop codons predicted to induce NMD. These alternative outcomes can lead to the ribosome translating downstream of such stop codons and hence the transcript escaping NMD. NMD escape via these mechanisms can have wide-ranging implications on human health, from being exploited by viruses to hijack host cell systems to being harnessed as potential therapeutic possibilities to treat genetic diseases.
Keywords: nonsense mutations; nonsense-meditated mRNA decay; ribosome; translation; translation termination.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

