Disentangling autoproteolytic cleavage from tethered agonist-dependent activation of the adhesion receptor ADGRL3
- PMID: 36244455
- PMCID: PMC9674912
- DOI: 10.1016/j.jbc.2022.102594
Disentangling autoproteolytic cleavage from tethered agonist-dependent activation of the adhesion receptor ADGRL3
Abstract
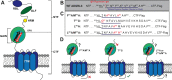
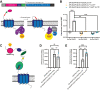
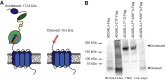
Adhesion G protein-coupled receptor latrophilin 3 (ADGRL3), a cell adhesion molecule highly expressed in the central nervous system, acts in synapse formation through trans interactions with its ligands. It is largely unknown if these interactions serve a purely adhesive function or can modulate G protein signaling. To assess how different structural elements of ADGRL3 (e.g., the adhesive domains, autoproteolytic cleavage site, or tethered agonist (TA)) impact receptor function, we require constructs that disrupt specific receptor features without impacting others. While we showed previously that mutating conserved Phe and Met residues in the TA of ADGRL3-C-terminal fragment (CTF), a CTF truncated to the G protein-coupled receptor proteolysis site, abolishes receptor-mediated G protein activation, we now find that autoproteolytic cleavage is disrupted in the full-length version of this construct. To identify a construct that disrupts TA-dependent activity without impacting proteolysis, we explored other mutations in the TA. We found that mutating the sixth and seventh residues of the TA, Leu and Met, to Ala impaired activity in a serum response element activity assay for both full-length and CTF constructs. We confirmed this activity loss results from impaired G protein coupling using an assay that acutely exposes the TA through controlled proteolysis. The ADGRL3 mutant expresses normally at the cell surface, and immunoblotting shows that it undergoes normal autoproteolysis. Thus, we found a construct that disrupts tethered agonism while retaining autoproteolytic cleavage, providing a tool to disentangle these functions in vivo. Our approach and specific findings are likely to be broadly applicable to other adhesion receptors.
Keywords: G protein; adhesion G protein-coupled receptor latrophilin; adhesion G protein-coupled receptors; autoproteolysis; cell signaling; membrane protein; tethered agonist.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

