Deregulated transcription factors in cancer cell metabolisms and reprogramming
- PMID: 36244530
- PMCID: PMC11220368
- DOI: 10.1016/j.semcancer.2022.10.001
Deregulated transcription factors in cancer cell metabolisms and reprogramming
Abstract
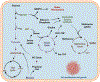
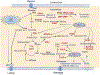
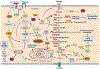
Metabolic reprogramming is an important cancer hallmark that plays a key role in cancer malignancies and therapy resistance. Cancer cells reprogram the metabolic pathways to generate not only energy and building blocks but also produce numerous key signaling metabolites to impact signaling and epigenetic/transcriptional regulation for cancer cell proliferation and survival. A deeper understanding of the mechanisms by which metabolic reprogramming is regulated in cancer may provide potential new strategies for cancer targeting. Recent studies suggest that deregulated transcription factors have been observed in various human cancers and significantly impact metabolism and signaling in cancer. In this review, we highlight the key transcription factors that are involved in metabolic control, dissect the crosstalk between signaling and transcription factors in metabolic reprogramming, and offer therapeutic strategies targeting deregulated transcription factors for cancer treatment.
Keywords: Cancer treatment; Cell metabolism; Metabolic reprogramming; Signaling metabolites; Transcription factors.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest
H.K.L. is a consultant for Stablix, Inc. All other authors declare no competing interests.
Figures


Similar articles
-
Crosstalk between metabolic reprogramming and epigenetics in cancer: updates on mechanisms and therapeutic opportunities.Cancer Commun (Lond). 2022 Nov;42(11):1049-1082. doi: 10.1002/cac2.12374. Epub 2022 Oct 20. Cancer Commun (Lond). 2022. PMID: 36266736 Free PMC article. Review.
-
Targeting Lipid Metabolic Reprogramming as Anticancer Therapeutics.J Cancer Prev. 2016 Dec;21(4):209-215. doi: 10.15430/JCP.2016.21.4.209. Epub 2016 Dec 30. J Cancer Prev. 2016. PMID: 28053954 Free PMC article. Review.
-
The role of ubiquitination and deubiquitination in cancer metabolism.Mol Cancer. 2020 Oct 1;19(1):146. doi: 10.1186/s12943-020-01262-x. Mol Cancer. 2020. PMID: 33004065 Free PMC article. Review.
-
Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression.Cell Mol Life Sci. 2016 Jan;73(2):377-92. doi: 10.1007/s00018-015-2070-4. Epub 2015 Oct 23. Cell Mol Life Sci. 2016. PMID: 26499846 Free PMC article. Review.
-
Cancer cell metabolism connects epigenetic modifications to transcriptional regulation.FEBS J. 2022 Mar;289(5):1302-1314. doi: 10.1111/febs.16032. Epub 2021 Jun 11. FEBS J. 2022. PMID: 34036737 Free PMC article. Review.
Cited by
-
Jun Dimerization Protein 2 (JDP2) Increases p53 Transactivation by Decreasing MDM2.Cancers (Basel). 2024 Feb 29;16(5):1000. doi: 10.3390/cancers16051000. Cancers (Basel). 2024. PMID: 38473360 Free PMC article.
-
The role of IKZF1 rs4132601 and Δ4-7 somatic deletion in acute lymphoblastic leukemia: a bioinformatics and case-control study.Mol Biol Rep. 2025 May 22;52(1):487. doi: 10.1007/s11033-025-10608-x. Mol Biol Rep. 2025. PMID: 40402344
-
ELK4 induced upregulation of HOMER3 promotes the proliferation and metastasis in glioma via Wnt/β-catenin/EMT signaling pathway.Biol Direct. 2025 Apr 9;20(1):48. doi: 10.1186/s13062-025-00643-w. Biol Direct. 2025. PMID: 40205485 Free PMC article.
-
Fumarate hydratase in cancer research: scientific trends and findings over 22 years.Discov Oncol. 2025 May 29;16(1):949. doi: 10.1007/s12672-025-02312-w. Discov Oncol. 2025. PMID: 40442547 Free PMC article.
-
Zinc finger protein 468 up-regulation of TFAM contributes to the malignant growth and cisplatin resistance of breast cancer cells.Cell Div. 2024 Mar 1;19(1):8. doi: 10.1186/s13008-024-00113-1. Cell Div. 2024. PMID: 38429817 Free PMC article.
References
-
- Weidemuller P, Kholmatov M, Petsalaki E, Zaugg JB, Transcription factors: bridge between cell signaling and gene regulation, Proteomics 21 (2021), e2000034. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

