Ex vivo engineered human plasma cells exhibit robust protein secretion and long-term engraftment in vivo
- PMID: 36245034
- PMCID: PMC9573882
- DOI: 10.1038/s41467-022-33787-8
Ex vivo engineered human plasma cells exhibit robust protein secretion and long-term engraftment in vivo
Abstract
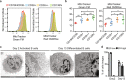
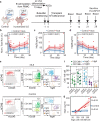
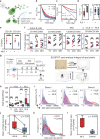
Due to their unique longevity and capacity to secrete high levels of protein, plasma B cells have the potential to be used as a cell therapy for protein replacement. Here, we show that ex vivo engineered human plasma cells exhibit single-cell RNA profiles, scanning electron micrograph ultrastructural features, and in vivo homing capacity of long-lived plasma cells. After transferring human plasma cells to immunodeficient mice in the presence of the human cytokines BAFF and IL-6, we observe increases in retention of plasma cells in the bone marrow, with engraftment exceeding a year. The most profound in vivo effects of human IL-6 are observed within 20 days of transfer and could be explained by decreased apoptosis in newly differentiated plasma cells. Collectively, these results show that ex vivo engineered and differentiated human plasma cells have the potential for long-lived in vivo protein secretion, which can be modeled in small animals.
© 2022. The Author(s).
Conflict of interest statement
A patent application has been filed (K.L.H., I.F.K., D.J.R., and R.G.J. “Engraftable cell-based immunotherapy for long-term delivery of therapeutic proteins,” US patent application no. US20180282692A1). The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

