Insights into the global freshwater virome
- PMID: 36246212
- PMCID: PMC9554406
- DOI: 10.3389/fmicb.2022.953500
Insights into the global freshwater virome
Abstract
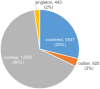
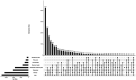
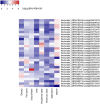
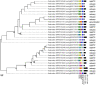
Viruses are by far the most abundant life forms on this planet. Yet, the full viral diversity remains mostly unknown, especially in environments like freshwater. Therefore, we aimed to study freshwater viruses in a global context. To this end, we downloaded 380 publicly available viral metagenomes (>1 TB). More than 60% of these metagenomes were discarded based on their levels of cellular contamination assessed by ribosomal DNA content. For the remaining metagenomes, assembled contigs were decontaminated using two consecutive steps, eventually yielding 273,365 viral contigs longer than 1,000 bp. Long enough contigs (≥ 10 kb) were clustered to identify novel genomes/genome fragments. We could recover 549 complete circular and high-quality draft genomes, out of which 10 were recognized as being novel. Functional annotation of these genomes showed that most of the annotated coding sequences are DNA metabolic genes or phage structural genes. On the other hand, taxonomic analysis of viral contigs showed that most of the assigned contigs belonged to the order Caudovirales, particularly the families of Siphoviridae, Myoviridae, and Podoviridae. The recovered viral contigs contained several auxiliary metabolic genes belonging to several metabolic pathways, especially carbohydrate and amino acid metabolism in addition to photosynthesis as well as hydrocarbon degradation and antibiotic resistance. Overall, we present here a set of prudently chosen viral contigs, which should not only help better understanding of freshwater viruses but also be a valuable resource for future virome studies.
Keywords: auxiliary metabolic genes; bacteriophages; freshwater; metagenome; virome.
Copyright © 2022 Elbehery and Deng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Extraordinary diversity of viruses in deep-sea sediments as revealed by metagenomics without prior virion separation.Environ Microbiol. 2021 Feb;23(2):728-743. doi: 10.1111/1462-2920.15154. Epub 2020 Aug 3. Environ Microbiol. 2021. PMID: 32627268
-
Viral metagenomes of Lake Soyang, the largest freshwater lake in South Korea.Sci Data. 2020 Oct 13;7(1):349. doi: 10.1038/s41597-020-00695-9. Sci Data. 2020. PMID: 33051444 Free PMC article.
-
Long-Read Metagenomics Improves the Recovery of Viral Diversity from Complex Natural Marine Samples.mSystems. 2022 Jun 28;7(3):e0019222. doi: 10.1128/msystems.00192-22. Epub 2022 Jun 13. mSystems. 2022. PMID: 35695508 Free PMC article.
-
Finer-Scale Phylosymbiosis: Insights from Insect Viromes.mSystems. 2018 Dec 18;3(6):e00131-18. doi: 10.1128/mSystems.00131-18. eCollection 2018 Nov-Dec. mSystems. 2018. PMID: 30574559 Free PMC article.
-
Metaviromics coupled with phage-host identification to open the viral 'black box'.J Microbiol. 2021 Mar;59(3):311-323. doi: 10.1007/s12275-021-1016-9. Epub 2021 Feb 23. J Microbiol. 2021. PMID: 33624268 Review.
Cited by
-
A Novel Tiled Amplicon Sequencing Assay Targeting the Tomato Brown Rugose Fruit Virus (ToBRFV) Genome Reveals Widespread Distribution in Municipal Wastewater Treatment Systems in the Province of Ontario, Canada.Viruses. 2024 Mar 17;16(3):460. doi: 10.3390/v16030460. Viruses. 2024. PMID: 38543825 Free PMC article.
-
Community Structure, Drivers, and Potential Functions of Different Lifestyle Viruses in Chaohu Lake.Viruses. 2024 Apr 11;16(4):590. doi: 10.3390/v16040590. Viruses. 2024. PMID: 38675931 Free PMC article.
-
Benchmarking informatics approaches for virus discovery: caution is needed when combining in silico identification methods.mSystems. 2024 Mar 19;9(3):e0110523. doi: 10.1128/msystems.01105-23. Epub 2024 Feb 20. mSystems. 2024. PMID: 38376167 Free PMC article.
-
Genomic insights reveal community structure and phylogenetic associations of endohyphal bacteria and viruses in fungal endophytes.Environ Microbiome. 2025 Jul 25;20(1):95. doi: 10.1186/s40793-025-00757-8. Environ Microbiome. 2025. PMID: 40713930 Free PMC article.
-
Genomic Analysis and Taxonomic Characterization of Seven Bacteriophage Genomes Metagenomic-Assembled from the Dishui Lake.Viruses. 2023 Sep 30;15(10):2038. doi: 10.3390/v15102038. Viruses. 2023. PMID: 37896815 Free PMC article.
References
-
- Auguet J. C., Montanié H., Hartmann H. J., Lebaron P., Casamayor E. O., Catala P., et al. (2009). Potential effect of freshwater virus on the structure and activity of bacterial communities in the Marennes-Oléron Bay (France). Microb. Ecol. 57, 295–306. doi: 10.1007/s00248-008-9428-1, PMID: - DOI - PubMed
-
- Bowman J. (2006). “The Methanotrophs — The families Methylococcaceae and Methylocystaceae,” in The Prokaryotes: Volume 5: Proteobacteria: Alpha and Beta Subclasses. eds. Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York, NY: Springer New York; ), 266–289.
LinkOut - more resources
Full Text Sources

