A new CCCH-type zinc finger-related lncRNA signature predicts the prognosis of clear cell renal cell carcinoma patients
- PMID: 36246657
- PMCID: PMC9562972
- DOI: 10.3389/fgene.2022.1034567
A new CCCH-type zinc finger-related lncRNA signature predicts the prognosis of clear cell renal cell carcinoma patients
Abstract
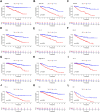
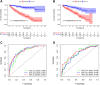
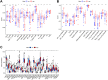
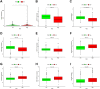
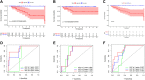
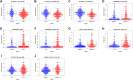
Background: Clear cell renal cell carcinoma (ccRCC) is the main component of renal cell carcinoma (RCC), and advanced ccRCC frequently indicates a poor prognosis. The significance of the CCCH-type zinc finger (CTZF) gene in cancer has been increasingly demonstrated during the past few years. According to studies, targeted radical therapy for cancer treatment may be a revolutionary therapeutic approach. Both lncRNAs and CCCH-type zinc finger genes are essential in ccRCC. However, the predictive role of long non-coding RNA (lncRNA) associated with the CCCH-type zinc finger gene in ccRCC needs further elucidation. This study aims to predict patient prognosis and investigate the immunological profile of ccRCC patients using CCCH-type zinc finger-associated lncRNAs (CTZFLs). Methods: From the Cancer Genome Atlas database, RNA-seq and corresponding clinical and prognostic data of ccRCC patients were downloaded. Univariate and multivariate Cox regression analyses were conducted to acquire CTZFLs for constructing prediction models. The risk model was verified using receiver operating characteristic curve analysis. The Kaplan-Meier method was used to analyze the overall survival (OS) of high-risk and low-risk groups. Multivariate Cox and stratified analyses were used to assess the prognostic value of the predictive feature in the entire cohort and different subgroups. In addition, the relationship between risk scores, immunological status, and treatment response was studied. Results: We constructed a signature consisting of eight CTZFLs (LINC02100, AC002451.1, DBH-AS1, AC105105.3, AL357140.2, LINC00460, DLGAP1-AS2, AL162377.1). The results demonstrated that the prognosis of ccRCC patients was independently predicted by CTZFLs signature and that the prognosis of high-risk groups was poorer than that of the lower group. CTZFLs markers had the highest diagnostic adequacy compared to single clinicopathologic factors, and their AUC (area under the receiver operating characteristic curve) was 0.806. The overall survival of high-risk groups was shorter than that of low-risk groups when patients were divided into groups based on several clinicopathologic factors. There were substantial differences in immunological function, immune cell score, and immune checkpoint expression between high- and low-risk groups. Additionally, Four agents, including ABT737, WIKI4, afuresertib, and GNE 317, were more sensitive in the high-risk group. Conclusion: The Eight-CTZFLs prognostic signature may be a helpful prognostic indicator and may help with medication selection for clear cell renal cell carcinoma.
Keywords: CCCH-type zinc finger; clear cell renal cell carcinoma; drug therapy; immune infiltration; lncRNAs.
Copyright © 2022 Shen, Chen, Jiang, Zhang, Xu, Peng, Zuo, Jiang, Fan, Fang and Zheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
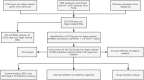
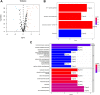

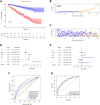
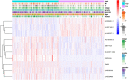


Similar articles
-
Identification of a C2H2 zinc finger-related lncRNA prognostic signature and its association with the immune microenvironment in clear cell renal cell carcinoma.Transl Androl Urol. 2025 Feb 28;14(2):412-431. doi: 10.21037/tau-2024-769. Epub 2025 Feb 25. Transl Androl Urol. 2025. PMID: 40114819 Free PMC article.
-
Computational construction of TME-related lncRNAs signature for predicting prognosis and immunotherapy response in clear cell renal cell carcinoma.J Clin Lab Anal. 2022 Aug;36(8):e24582. doi: 10.1002/jcla.24582. Epub 2022 Jul 8. J Clin Lab Anal. 2022. PMID: 35808868 Free PMC article.
-
Development of a novel disulfidptosis-related lncRNA signature for prognostic and immune response prediction in clear cell renal cell carcinoma.Sci Rep. 2024 Jan 5;14(1):624. doi: 10.1038/s41598-024-51197-2. Sci Rep. 2024. PMID: 38182642 Free PMC article.
-
Current Concepts of Non-Coding RNAs in the Pathogenesis of Non-Clear Cell Renal Cell Carcinoma.Cancers (Basel). 2019 Oct 17;11(10):1580. doi: 10.3390/cancers11101580. Cancers (Basel). 2019. PMID: 31627266 Free PMC article. Review.
-
Review of Prognostic Expression Markers for Clear Cell Renal Cell Carcinoma.Front Oncol. 2021 Apr 28;11:643065. doi: 10.3389/fonc.2021.643065. eCollection 2021. Front Oncol. 2021. PMID: 33996558 Free PMC article. Review.
Cited by
-
Identification of a lactate metabolism-related lncRNAs signature for predicting the prognosis in patients with kidney renal clear cell carcinoma.Transl Androl Urol. 2024 Apr 30;13(4):509-525. doi: 10.21037/tau-23-483. Epub 2024 Apr 18. Transl Androl Urol. 2024. PMID: 38721281 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

