Computational approaches for predicting variant impact: An overview from resources, principles to applications
- PMID: 36246661
- PMCID: PMC9559863
- DOI: 10.3389/fgene.2022.981005
Computational approaches for predicting variant impact: An overview from resources, principles to applications
Abstract
One objective of human genetics is to unveil the variants that contribute to human diseases. With the rapid development and wide use of next-generation sequencing (NGS), massive genomic sequence data have been created, making personal genetic information available. Conventional experimental evidence is critical in establishing the relationship between sequence variants and phenotype but with low efficiency. Due to the lack of comprehensive databases and resources which present clinical and experimental evidence on genotype-phenotype relationship, as well as accumulating variants found from NGS, different computational tools that can predict the impact of the variants on phenotype have been greatly developed to bridge the gap. In this review, we present a brief introduction and discussion about the computational approaches for variant impact prediction. Following an innovative manner, we mainly focus on approaches for non-synonymous variants (nsSNVs) impact prediction and categorize them into six classes. Their underlying rationale and constraints, together with the concerns and remedies raised from comparative studies are discussed. We also present how the predictive approaches employed in different research. Although diverse constraints exist, the computational predictive approaches are indispensable in exploring genotype-phenotype relationship.
Keywords: genotype-phenotype relationship; human genetics; in silico prediction; nonsynonymous variants; variant impact.
Copyright © 2022 Liu, Yeung, Chiu and Cao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
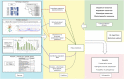
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

