Genetic analysis of seven pateints with Hereditary Multiple Osteochondromas (HMO)
- PMID: 36247276
- PMCID: PMC9556467
Genetic analysis of seven pateints with Hereditary Multiple Osteochondromas (HMO)
Abstract
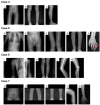
Background: HMO (Hereditary Multiple Osteochondroma), an uncommon autosomal dominant disorder, is characterized by the development of multiple osteochondromas, which are nonmalignant cartilage-capped bone tumors growing outwards from long bone metaphyses.
Methods: The present work retrospectively analyzed seven children with HMO who were enrolled for routine clinical diagnosis and treatment, including X-ray examination. Subsequent genetic detection was carried out using whole exome sequencing (WES). In addition, this work applied Sanger sequencing to be the validation approach. Moreover, this work also examined amino acid (AA) evolutionary conservatism under the influence of certain missense variants.
Results: The clinical indications of all seven patients and their family members were thoroughly indexed. WES identified diagnostic variants in the EXT1 or EXT2 gene in these patients. In these variants, four were reported for the first time, namely EXT1: c.1285-2A>T, EXT2: c.1139delT, EXT1: c.203G>A, and EXT1: c.1645_1673del. Familial validation revealed that three of the variants were hereditary, while the other four were de novo, which was consistent with the phenotype in each case.
Conclusion: Our results expanded HMO variation spectrum, and laid certain foundations for the precise counseling of those affected families.
Keywords: EXT1 gene; EXT2 gene; Hereditary multiple osteochondromas; Novel mutation; Whole-exome sequencing.
AJTR Copyright © 2022.
Conflict of interest statement
None.
Figures


References
-
- Wuyts W, Schmale GA, Chansky HA, Raskind WH. Hereditary Multiple Osteochondromas. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle; 1993-2022. 2000 Aug 3 [Updated 2020 Aug 6] - PubMed
-
- Ryckx A, Somers JF, Allaert L. Hereditary multiple exostosis. Acta Orthop Belg. 2013;79:597–607. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
