Genome-wide association, RNA-seq and iTRAQ analyses identify candidate genes controlling radicle length of wheat
- PMID: 36247556
- PMCID: PMC9554269
- DOI: 10.3389/fpls.2022.939544
Genome-wide association, RNA-seq and iTRAQ analyses identify candidate genes controlling radicle length of wheat
Abstract
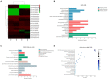
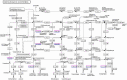
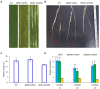
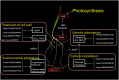
The radicle, present in the embryo of a seed, is the first root to emerge at germination, and its rapid growth is essential for establishment and survival of the seedling. However, there are few studies on the critical mechanisms underlying radicle and then radicle length in wheat seedlings, despite its importance as a food crop throughout the world. In the present study, 196 wheat accessions from the Huanghuai Wheat Region were screened to measure radicle length under 4 hydroponic culture environments over 3 years. Different expression genes and proteins (DEGs/DEPs) between accessions with extremely long [Yunong 949 (WRL1), Zhongyu 9,302 (WRL2)] and short roots [Yunong 201 (WRS1), Beijing 841 (WRS2)] were identified in 12 sets of root tissue samples by RNA-seq and iTRAQ (Isobaric tags for relative and absolute quantification). Phenotypic results showed that the elongation zone was significantly longer in root accessions with long roots compared to the short-rooted accessions. A genome-wide association study (GWAS) identified four stable chromosomal regions significantly associated with radicle length, among which 1A, 4A, and 7A chromosomes regions explained 7.17% to12.93% of the phenotypic variation. The omics studies identified the expression patterns of 24 DEGs/DEPs changed at both the transcriptional and protein levels. These DEGs/DEPs were mainly involved in carbon fixation in photosynthetic organisms, photosynthesis and phenylpropanoid biosynthesis pathways. TraesCS1A02G104100 and TraesCS2B02G519100 were involved in the biosynthesis of tricin-lignins in cell walls and may affect the extension of cell walls in the radicle elongation zone. A combination of GWAS and RNA-seq analyses revealed 19 DEGs with expression changes in the four accessions, among which, TraesCS1A02G422700 (a cysteine-rich receptor-like protein kinase 6, CRK6) also showed upregulation in the comparison group by RNA-seq, iTRAQ, and qRT-PCR. BSMV-mediated gene silencing also showed that TaCRK6 improves root development in wheat. Our data suggest that TaCRK6 is a candidate gene regulating radicle length in wheat.
Keywords: GWAS analysis; RNA-seq analysis; iTRAQ analysis; radicle length; wheat (Triticum aestivum L.).
Copyright © 2022 Xu, Chen, Zhou, Yue, Yang, Zhang, Zhan and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188. doi: 10.1214/aos/1013699998 - DOI
-
- Biscarini F., Cozzi P., Casella L., Riccardi P., Vattari A., Orasen G., et al. . (2016). Genome-wide association study for traits related to plant and grain morphology, and root architecture in temperate rice accessions. PLoS One 11:e0155425. doi: 10.1371/journal.pone.0155425, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

