Integrating metabolite and transcriptome analysis revealed the different mechanisms of characteristic compound biosynthesis and transcriptional regulation in tea flowers
- PMID: 36247612
- PMCID: PMC9557745
- DOI: 10.3389/fpls.2022.1016692
Integrating metabolite and transcriptome analysis revealed the different mechanisms of characteristic compound biosynthesis and transcriptional regulation in tea flowers
Abstract
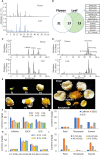
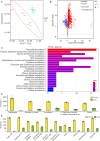
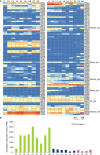
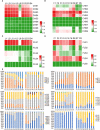
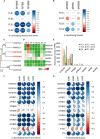
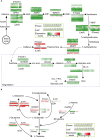
The flowers of tea plants (Camellia sinensis), as well as tea leaves, contain abundant secondary metabolites and are big potential resources for the extraction of bioactive compounds or preparation of functional foods. However, little is known about the biosynthesis and transcriptional regulation mechanisms of those metabolites in tea flowers, such as terpenoid, flavonol, catechins, caffeine, and theanine. This study finely integrated target and nontarget metabolism analyses to explore the metabolic feature of developing tea flowers. Tea flowers accumulated more abundant terpenoid compounds than young leaves. The transcriptome data of developing flowers and leaves showed that a higher expression level of later genes of terpenoid biosynthesis pathway, such as Terpene synthases gene family, in tea flowers was the candidate reason of the more abundant terpenoid compounds than in tea leaves. Differently, even though flavonol and catechin profiling between tea flowers and leaves was similar, the gene family members of flavonoid biosynthesis were selectively expressed by tea flowers and tea leaves. Transcriptome and phylogenetic analyses indicated that the regulatory mechanism of flavonol biosynthesis was perhaps different between tea flowers and leaves. However, the regulatory mechanism of catechin biosynthesis was perhaps similar between tea flowers and leaves. This study not only provides a global vision of metabolism and transcriptome in tea flowers but also uncovered the different mechanisms of biosynthesis and transcriptional regulation of those important compounds.
Keywords: metabolism; regulation mechanism; tea flowers; transcription factor; transcriptome.
Copyright © 2022 Tang, Shen, Li, Yue, Duan, Ye, Lin, Zhou, Yang, Chen, Wang, Zhao and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Global transcriptome and gene regulation network for secondary metabolite biosynthesis of tea plant (Camellia sinensis).BMC Genomics. 2015 Jul 29;16(1):560. doi: 10.1186/s12864-015-1773-0. BMC Genomics. 2015. PMID: 26220550 Free PMC article.
-
Transcriptomic and phytochemical analysis of the biosynthesis of characteristic constituents in tea (Camellia sinensis) compared with oil tea (Camellia oleifera).BMC Plant Biol. 2015 Aug 7;15:190. doi: 10.1186/s12870-015-0574-6. BMC Plant Biol. 2015. PMID: 26245644 Free PMC article.
-
Integrated transcriptomics and metabolomics analysis of catechins, caffeine and theanine biosynthesis in tea plant (Camellia sinensis) over the course of seasons.BMC Plant Biol. 2020 Jun 29;20(1):294. doi: 10.1186/s12870-020-02443-y. BMC Plant Biol. 2020. PMID: 32600265 Free PMC article.
-
Exploring plant metabolic genomics: chemical diversity, metabolic complexity in the biosynthesis and transport of specialized metabolites with the tea plant as a model.Crit Rev Biotechnol. 2020 Aug;40(5):667-688. doi: 10.1080/07388551.2020.1752617. Epub 2020 Apr 22. Crit Rev Biotechnol. 2020. PMID: 32321331 Review.
-
Occurrence of Functional Molecules in the Flowers of Tea (Camellia sinensis) Plants: Evidence for a Second Resource.Molecules. 2018 Mar 29;23(4):790. doi: 10.3390/molecules23040790. Molecules. 2018. PMID: 29596355 Free PMC article. Review.
Cited by
-
Unravelling the microbiome of wild flowering plants: a comparative study of leaves and flowers in alpine ecosystems.BMC Microbiol. 2024 Oct 19;24(1):417. doi: 10.1186/s12866-024-03574-0. BMC Microbiol. 2024. PMID: 39425049 Free PMC article.
-
Divergent MYB paralogs determine spatial distribution of linalool mediated by JA and DNA demethylation participating in aroma formation and cold tolerance of tea plants.Plant Biotechnol J. 2025 May;23(5):1455-1475. doi: 10.1111/pbi.14598. Epub 2025 Feb 11. Plant Biotechnol J. 2025. PMID: 39932489 Free PMC article.
-
Metabolite Profiling of External and Internal Petals in Three Different Colors of Tea Flowers (Camellia sinensis) Using Widely Targeted Metabolomics.Metabolites. 2023 Jun 23;13(7):784. doi: 10.3390/metabo13070784. Metabolites. 2023. PMID: 37512491 Free PMC article.
-
Widely Targeted Metabolomics Analysis of the Roots, Stems, Leaves, Flowers, and Fruits of Camellia luteoflora, a Species with an Extremely Small Population.Molecules. 2024 Oct 8;29(19):4754. doi: 10.3390/molecules29194754. Molecules. 2024. PMID: 39407682 Free PMC article.
-
Deciphering aroma formation during flowering in nectar tree (Tilia amurensis): insights from integrated metabolome and transcriptome analysis.For Res (Fayettev). 2023 Oct 8;3:24. doi: 10.48130/FR-2023-0024. eCollection 2023. For Res (Fayettev). 2023. PMID: 39526254 Free PMC article.
References
LinkOut - more resources
Full Text Sources

