Integrated analyses of transcriptome and metabolome provides new insights into the primary and secondary metabolism in response to nitrogen deficiency and soil compaction stress in peanut roots
- PMID: 36247623
- PMCID: PMC9554563
- DOI: 10.3389/fpls.2022.948742
Integrated analyses of transcriptome and metabolome provides new insights into the primary and secondary metabolism in response to nitrogen deficiency and soil compaction stress in peanut roots
Abstract
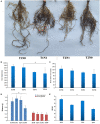
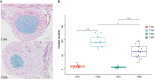
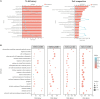
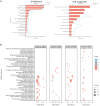
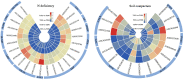
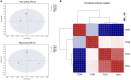
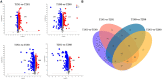
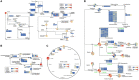
Peanut (Arachis hypogaea L.) is an important oil crop globally because of its high edible and economic value. However, its yield and quality are often restricted by certain soil factors, especially nitrogen (N) deficiency, and soil compaction. To explore the molecular mechanisms and metabolic basis behind the peanut response to N deficiency and soil compaction stresses, transcriptome and metabolome analyses of peanut root were carried out. The results showed that N deficiency and soil compaction stresses clearly impaired the growth and development of peanut's aboveground and underground parts, as well as its root nodulation. A total of 18645 differentially expressed genes (DEGs) and 875 known differentially accumulated metabolites (DAMs) were identified in peanut root under differing soil compaction and N conditions. The transcriptome analysis revealed that DEGs related to N deficiency were mainly enriched in "amino acid metabolism", "starch and sucrose metabolism", and "TCA cycle" pathways, while DEGs related to soil compaction were mainly enriched in "oxidoreductase activity", "lipids metabolism", and "isoflavonoid biosynthesis" pathways. The metabolome analysis also showed significant differences in the accumulation of metabolisms in these pathways under different stress conditions. Then the involvement of genes and metabolites in pathways of "amino acid metabolism", "TCA cycle", "lipids metabolism", and "isoflavonoid biosynthesis" under different soil compaction and N deficiency stresses were well discussed. This integrated transcriptome and metabolome analysis study enhances our mechanistic knowledge of how peanut plants respond to N deficiency and soil compaction stresses. Moreover, it provides new leads to further investigate candidate functional genes and metabolic pathways for use in improving the adaptability of peanut to abiotic stress and accelerating its breeding process of new stress-resistant varieties.
Keywords: metabolome; nitrogen deficiency; peanut root; soil compaction stress; transcriptome.
Copyright © 2022 Yang, Wu, Liang, Yin and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Comprehensive Transcriptome and Metabolome Analyses Reveal Primary Molecular Regulation Pathways Involved in Peanut under Water and Nitrogen Co-Limitation.Int J Mol Sci. 2023 Aug 27;24(17):13308. doi: 10.3390/ijms241713308. Int J Mol Sci. 2023. PMID: 37686113 Free PMC article.
-
Integrated analyses reveal the response of peanut to phosphorus deficiency on phenotype, transcriptome and metabolome.BMC Plant Biol. 2022 Nov 14;22(1):524. doi: 10.1186/s12870-022-03867-4. BMC Plant Biol. 2022. PMID: 36372886 Free PMC article.
-
Integrative Analysis of Metabolome and Transcriptome Reveals Molecular Mechanisms Regulating Oil and Protein Content in Peanut (Arachis hypogaea L).J Agric Food Chem. 2024 Nov 27;72(47):26586-26598. doi: 10.1021/acs.jafc.4c07999. Epub 2024 Nov 13. J Agric Food Chem. 2024. PMID: 39539052
-
The key metabolic pathway of roots and leaves responses in Arachis hypogaea under Al toxicity stress.BMC Plant Biol. 2025 Apr 7;25(1):439. doi: 10.1186/s12870-025-06460-7. BMC Plant Biol. 2025. PMID: 40189501 Free PMC article.
-
The genus Arachis: an excellent resource for studies on differential gene expression for stress tolerance.Front Plant Sci. 2023 Oct 30;14:1275854. doi: 10.3389/fpls.2023.1275854. eCollection 2023. Front Plant Sci. 2023. PMID: 38023864 Free PMC article. Review.
Cited by
-
Integrated transcriptomic and metabolomic analyses reveal key metabolic pathways in response to potassium deficiency in coconut (Cocos nucifera L.) seedlings.Front Plant Sci. 2023 Feb 13;14:1112264. doi: 10.3389/fpls.2023.1112264. eCollection 2023. Front Plant Sci. 2023. PMID: 36860901 Free PMC article.
-
Comprehensive Transcriptome and Metabolome Analyses Reveal Primary Molecular Regulation Pathways Involved in Peanut under Water and Nitrogen Co-Limitation.Int J Mol Sci. 2023 Aug 27;24(17):13308. doi: 10.3390/ijms241713308. Int J Mol Sci. 2023. PMID: 37686113 Free PMC article.
-
Multi-year crop rotation and quicklime application promote stable peanut yield and high nutrient-use efficiency by regulating soil nutrient availability and bacterial/fungal community.Front Microbiol. 2024 May 17;15:1367184. doi: 10.3389/fmicb.2024.1367184. eCollection 2024. Front Microbiol. 2024. PMID: 38827150 Free PMC article.
-
Alfalfa Responses to Intensive Soil Compaction: Effects on Plant and Root Growth, Phytohormones and Internal Gene Expression.Plants (Basel). 2024 Mar 26;13(7):953. doi: 10.3390/plants13070953. Plants (Basel). 2024. PMID: 38611482 Free PMC article.
References
-
- Abbott L. K., Murphy D. V. (2007). Soil Biological Fertility. A Key to Sustainable Land Use in Agriculture, Berlin: Springer. 10.1007/978-1-4020-6619-1 - DOI
-
- Arvidsson J. (1999). Nutrient uptake and growth of barley as affected by soil compaction. Plant Soil. 208, 9–19. 10.1023/A:1004484518652 - DOI
-
- Arvidsson J., Etana A., Rydberg T. (2014). Crop yield in Swedish experiments with shallow tillage and no-tillage 1983–2012. Eur. J. Agron. 52, 307–315. 10.1016/j.eja.2013.08.002 - DOI
LinkOut - more resources
Full Text Sources

